Emerging roles for R-loop structures in the management of topological stress
- PMID: 32107311
- PMCID: PMC7135976
- DOI: 10.1074/jbc.REV119.006364
Emerging roles for R-loop structures in the management of topological stress
Abstract
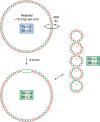
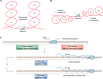
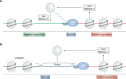
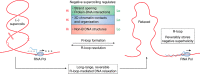
R-loop structures are a prevalent class of alternative non-B DNA structures that form during transcription upon invasion of the DNA template by the nascent RNA. R-loops form universally in the genomes of organisms ranging from bacteriophages, bacteria, and yeasts to plants and animals, including mammals. A growing body of work has linked these structures to both physiological and pathological processes, in particular to genome instability. The rising interest in R-loops is placing new emphasis on understanding the fundamental physicochemical forces driving their formation and stability. Pioneering work in Escherichia coli revealed that DNA topology, in particular negative DNA superhelicity, plays a key role in driving R-loops. A clear role for DNA sequence was later uncovered. Here, we review and synthesize available evidence on the roles of DNA sequence and DNA topology in controlling R-loop formation and stability. Factoring in recent developments in R-loop modeling and single-molecule profiling, we propose a coherent model accounting for the interplay between DNA sequence and DNA topology in driving R-loop structure formation. This model reveals R-loops in a new light as powerful and reversible topological stress relievers, an insight that significantly expands the repertoire of R-loops' potential biological roles under both normal and aberrant conditions.
Keywords: DNA structure; DNA topoisomerase; DNA topology; DNA transcription; R-loop; chromosomes; gene transcription; genome stability; supercoiling; superhelicity; topological stress.
© 2020 Chedin and Benham.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures