Molecular and genetic regulation of pig pancreatic islet cell development
- PMID: 32108026
- PMCID: PMC7132804
- DOI: 10.1242/dev.186213
Molecular and genetic regulation of pig pancreatic islet cell development
Abstract
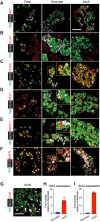
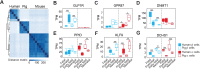
Reliance on rodents for understanding pancreatic genetics, development and islet function could limit progress in developing interventions for human diseases such as diabetes mellitus. Similarities of pancreas morphology and function suggest that porcine and human pancreas developmental biology may have useful homologies. However, little is known about pig pancreas development. To fill this knowledge gap, we investigated fetal and neonatal pig pancreas at multiple, crucial developmental stages using modern experimental approaches. Purification of islet β-, α- and δ-cells followed by transcriptome analysis (RNA-seq) and immunohistology identified cell- and stage-specific regulation, and revealed that pig and human islet cells share characteristic features that are not observed in mice. Morphometric analysis also revealed endocrine cell allocation and architectural similarities between pig and human islets. Our analysis unveiled scores of signaling pathways linked to native islet β-cell functional maturation, including evidence of fetal α-cell GLP-1 production and signaling to β-cells. Thus, the findings and resources detailed here show how pig pancreatic islet studies complement other systems for understanding the developmental programs that generate functional islet cells, and that are relevant to human pancreatic diseases.
Keywords: Diabetes mellitus; Metabolism; Organogenesis; Pancreas; Pig; α-Cell; β-Cell; δ-cell.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

