Ab Initio Construction and Evolutionary Analysis of Protein-Coding Gene Families with Partially Homologous Relationships: Closely Related Drosophila Genomes as a Case Study
- PMID: 32108239
- PMCID: PMC7144356
- DOI: 10.1093/gbe/evaa041
Ab Initio Construction and Evolutionary Analysis of Protein-Coding Gene Families with Partially Homologous Relationships: Closely Related Drosophila Genomes as a Case Study
Abstract
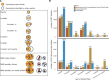
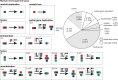
How have genes evolved within a well-known genome phylogeny? Many protein-coding genes should have evolved as a whole at the gene level, and some should have evolved partly through fragments at the subgene level. To comprehensively explore such complex homologous relationships and better understand gene family evolution, here, with de novo-identified modules, the subgene units which could consecutively cover proteins within a set of closely related species, we applied a new phylogeny-based approach that considers evolutionary models with partial homology to classify all protein-coding genes in nine Drosophila genomes. Compared with two other popular methods for gene family construction, our approach improved practical gene family classifications with a more reasonable view of homology and provided a much more complete landscape of gene family evolution at the gene and subgene levels. In the case study, we found that most expanded gene families might have evolved mainly through module rearrangements rather than gene duplications and mainly generated single-module genes through partial gene duplication, suggesting that there might be pervasive subgene rearrangement in the evolution of protein-coding gene families. The use of a phylogeny-based approach with partial homology to classify and analyze protein-coding gene families may provide us with a more comprehensive landscape depicting how genes evolve within a well-known genome phylogeny.
Keywords: evolution; gene family; module architecture; partial homology; subgene rearrangement.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Ane C, et al. 2006. Bayesian estimation of concordance among gene trees. Mol Biol Evol. 24(2):412–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

