Tumor Mutational Burden From Tumor-Only Sequencing Compared With Germline Subtraction From Paired Tumor and Normal Specimens
- PMID: 32108894
- PMCID: PMC7049088
- DOI: 10.1001/jamanetworkopen.2020.0202
Tumor Mutational Burden From Tumor-Only Sequencing Compared With Germline Subtraction From Paired Tumor and Normal Specimens
Abstract
Importance: Tumor mutation burden (TMB) is an emerging factor associated with survival with immunotherapy. When tumor-normal pairs are available, TMB is determined by calculating the difference between somatic and germline sequences. In the case of commonly used tumor-only sequencing, additional steps are needed to estimate the somatic alterations. Computational tools have been developed to determine germline contribution based on sample copy state, purity estimates, and occurrence of the variant in population databases; however, there is potential for sampling bias in population data sets.
Objective: To investigate whether tumor-only filtering approaches overestimate TMB.
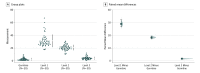
Design, setting, and participants: This was a retrospective cohort study of 50 tumor samples from 10 different tumor types. A 595-gene panel test was used to assess TMB by adding all missense, indels, and frameshift variants with an allelic fraction of at least 5% and coverage of at least 100× within each tumor. Tumor-only TMB was evaluated against the criterion standard of matched germline-subtracted TMB at 3 levels. Level 1 removed all the tumor-only variants with allelic fraction of at least 1% in the Exome Aggregation Consortium database (with the Cancer Genome Atlas cohort removed). Level 2 removed all variants observed in population databases, simulating a naive approach of removing germline variation. Level 3 used an internal tumor-only pipeline for calculating TMB. These specimens were processed with a commercially available panel, and results were analyzed at the Mayo Clinic. Data were analyzed between December 1, 2018, and May 28, 2019.
Main outcomes and measures: Tumor mutation burden per megabase (Mb) as determined by 3 levels of filtering and germline subtraction.
Results: There were significantly higher estimates of TMB with level 1 (median [range] mutations per Mb, 28.8 [17.5-67.1]), level 2 (median [range] mutations per Mb, 20.8 [10.4-30.8]), and level 3 (median [range] mutations per Mb, 3.8 [0.8-12.1]) tumor-only filtering approaches than those determined by germline subtraction (median [range] mutations per Mb, 1.7 [0.4-9.2]). There were no strong associations between TMB estimates and tumor-germline TMB for level 1 filtering (r = 0.008; 95% CI, -0.004 to 0.020), level 2 filtering (r = 0.018; 95% CI, 0.003 to 0.033), or level 3 filtering (r = 0.54; 95% CI, 0.36 to 0.68).
Conclusions and relevance: The findings of this study indicate that tumor-only approaches that filter variants in population databases can overestimate TMB compared with germline subtraction methods. Despite improved association with more stringent filtering approaches, these falsely elevated estimates may result in the inappropriate categorization of tumor specimens and negatively affect clinical trial results and patient outcomes.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

