A multi-omics dataset of heat-shock response in the yeast RNA binding protein Mip6
- PMID: 32109230
- PMCID: PMC7046740
- DOI: 10.1038/s41597-020-0412-z
A multi-omics dataset of heat-shock response in the yeast RNA binding protein Mip6
Abstract
Gene expression is a biological process regulated at different molecular levels, including chromatin accessibility, transcription, and RNA maturation and transport. In addition, these regulatory mechanisms have strong links with cellular metabolism. Here we present a multi-omics dataset that captures different aspects of this multi-layered process in yeast. We obtained RNA-seq, metabolomics, and H4K12ac ChIP-seq data for wild-type and mip6Δ strains during a heat-shock time course. Mip6 is an RNA-binding protein that contributes to RNA export during environmental stress and is informative of the contribution of post-transcriptional regulation to control cellular adaptations to environmental changes. The experiment was performed in quadruplicate, and the different omics measurements were obtained from the same biological samples, which facilitates the integration and analysis of data using covariance-based methods. We validate our dataset by showing that ChIP-seq, RNA-seq and metabolomics signals recapitulate existing knowledge about the response of ribosomal genes and the contribution of trehalose metabolism to heat stress. Raw data, processed data and preprocessing scripts are made available.
Conflict of interest statement
The authors declare no competing interests.
Figures
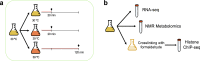




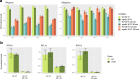


References
-
- García-Oliver E, García-Molinero V, Rodríguez-Navarro S. mRNA export and gene expression: The SAGA–TREX-2 connection. BBA-Gene Regul. Mech. 2012;1819:555–565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- PROMETEO/2016/093/Regional Government of Valencia | Conselleria d'Educació, Investigació, Cultura i Esport (Conselleria d'Educació, Investigació, Cultura i Esport de la Generalitat Valenciana)/International
- PROMETEO/2016/093/Regional Government of Valencia | Conselleria d'Educació, Investigació, Cultura i Esport (Conselleria d'Educació, Investigació, Cultura i Esport de la Generalitat Valenciana)/International
- PROMETEO/2016/093/Regional Government of Valencia | Conselleria d'Educació, Investigació, Cultura i Esport (Conselleria d'Educació, Investigació, Cultura i Esport de la Generalitat Valenciana)/International
- PROMETEO/2016/093/Regional Government of Valencia | Conselleria d'Educació, Investigació, Cultura i Esport (Conselleria d'Educació, Investigació, Cultura i Esport de la Generalitat Valenciana)/International
- PROMETEO/2016/093/Regional Government of Valencia | Conselleria d'Educació, Investigació, Cultura i Esport (Conselleria d'Educació, Investigació, Cultura i Esport de la Generalitat Valenciana)/International

