Long Non‑Coding RNAs Regulate Inflammation in Diabetic Peripheral Neuropathy by Acting as ceRNAs Targeting miR-146a-5p
- PMID: 32110074
- PMCID: PMC7035891
- DOI: 10.2147/DMSO.S242789
Long Non‑Coding RNAs Regulate Inflammation in Diabetic Peripheral Neuropathy by Acting as ceRNAs Targeting miR-146a-5p
Abstract
Background: Long non-coding RNAs (lncRNAs), as competing endogenous RNAs (ceRNAs), can regulate various pathophysiological processes by binding competitively to microRNAs at the post-transcription level. Our previous work demonstrated that miR-146a-5p was lowly expressed in diabetic peripheral neuropathy (DPN) rats. However, the ceRNA network in DPN mediated by lncRNAs and miR-146a-5p remains to be explored.
Methods: Two groups of rats (n=4 per group), a type 2 diabetes (T2DM) group and a DPN group, were used in this study. Sciatic nerve conduction velocity (NCV) of each rat was determined at the 6th and the 12th week. LncRNA microarray analysis was performed in the sciatic nerve of DPN and T2DM rats. Based on the TargetScan algorithm and the miRanda database, we determined the differentially expressed (DE) lncRNAs bound to miR-146a-5p. Furthermore, we verified the DE lncRNAs potentially bound to miR-146a-5p by qRT-PCR. The genes targeted by miR-146a-5p were identified by bioinformatics prediction and experimental techniques.
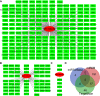
Results: We found 413 DE lncRNAs between DPN and T2DM rats (|log2FC| ≥ 2 and adjust P ≤ 0.05). Eight DE lncRNAs were predicted to bind to miR-146a-5p by both algorithms, of which four were verified by qRT-PCR. TRAF6, IRAK1, and SMAD4 were identified as miR-146a-5p targeted genes and were predominantly enriched in the inflammatory signaling pathway.
Conclusion: LncRNAs may contribute to the pathogenesis of DPN by regulating inflammation through functioning as ceRNAs of miR-146a-5p.
Keywords: MicroRNA; diabetic peripheral neuropathy; inflammation; long non-coding RNA.
© 2020 Feng et al.
Conflict of interest statement
The authors report no conflicts of interest for this work.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

