Integrated Mobile Element Scanning (ME-Scan) method for identifying multiple types of polymorphic mobile element insertions
- PMID: 32110248
- PMCID: PMC7035633
- DOI: 10.1186/s13100-020-00207-x
Integrated Mobile Element Scanning (ME-Scan) method for identifying multiple types of polymorphic mobile element insertions
Abstract
Background: Mobile elements are ubiquitous components of mammalian genomes and constitute more than half of the human genome. Polymorphic mobile element insertions (pMEIs) are a major source of human genomic variation and are gaining research interest because of their involvement in gene expression regulation, genome integrity, and disease.
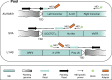
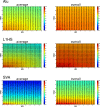
Results: Building on our previous Mobile Element Scanning (ME-Scan) protocols, we developed an integrated ME-Scan protocol to identify three major active families of human mobile elements, AluYb, L1HS, and SVA. This approach selectively amplifies insertion sites of currently active retrotransposons for Illumina sequencing. By pooling the libraries together, we can identify pMEIs from all three mobile element families in one sequencing run. To demonstrate the utility of the new ME-Scan protocol, we sequenced 12 human parent-offspring trios. Our results showed high sensitivity (> 90%) and accuracy (> 95%) of the protocol for identifying pMEIs in the human genome. In addition, we also tested the feasibility of identifying somatic insertions using the protocol.
Conclusions: The integrated ME-Scan protocol is a cost-effective way to identify novel pMEIs in the human genome. In addition, by developing the protocol to detect three mobile element families, we demonstrate the flexibility of the ME-Scan protocol. We present instructions for the library design, a sequencing protocol, and a computational pipeline for downstream analyses as a complete framework that will allow researchers to easily adapt the ME-Scan protocol to their own projects in other genomes.
Keywords: AluYb; High-throughput sequencing; LINE-1; ME-Scan; Mobile element insertion; Retrotransposon; SVA.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures

References
LinkOut - more resources
Full Text Sources

