Twists or turns: stabilising alpha vs. beta turns in tetrapeptides
- PMID: 32110345
- PMCID: PMC7020788
- DOI: 10.1039/c9sc04153b
Twists or turns: stabilising alpha vs. beta turns in tetrapeptides
Abstract
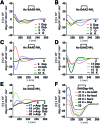
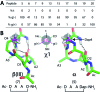
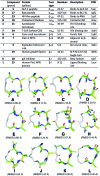
Protein-protein interactions involve hotspots as small as 4 sequential amino acids. Corresponding tetrapeptides have no structure in water. Here we report linking side chains of amino acids X and Z to form 24 cyclic tetrapeptides, cyclo-[XAAZ]-NH2, and stabilise 14-18 membered rings that mimic different kinds of non-regular secondary structures found in protein hotspots. 2D NMR spectra allowed determination of 3D structures for 14 cyclic tetrapeptides in water. Five formed two (i, i + 3) hydrogen bonds and a beta/gamma (6, 7) or beta (9, 19, 20) turn; eight formed one (i, i + 4) hydrogen bond and twisted into a non-helical (13, 18, 21, 22, 24) or helical (5, 17, 23) alpha turn; one was less structured (15). A beta or gamma turn was favoured for Z = Dab, Orn or Glu due to a χ1 gauche (+) rotamer, while an alpha turn was favoured for Z = Dap (but not X = Dap) due to a gauche (-) rotamer. Surprisingly, an unstructured peptide ARLARLARL could be twisted into a helix when either a helical or non-helical alpha turn (5, 13, 17, 18, 21-24) with Z = Dap was attached to the N-terminus. These structural models provide insights into stability for different turns and twists corresponding to non-regular folds in protein hotspots.
This journal is © The Royal Society of Chemistry 2019.
Figures


References
-
- Pavone V., Gaeta G., Lombardi A., Nastri F., Maglio O., Isernia C., Saviano M. Biopolymers. 1996;38:705–721. - PubMed
- Nataraj D. V., Srinivasan N., Sowdhamini R., Ramakrishnan C. Curr. Sci. 1995;69:434–447.
-
- Hoang H. N., Driver R. W., Beyer R. L., Malde A. K., Le G. T., Abbenante G., Mark A. E., Fairlie D. P. Angew. Chem., Int. Ed. 2011;50:11107–11111. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

