Translational Regulations in Response to Endoplasmic Reticulum Stress in Cancers
- PMID: 32111004
- PMCID: PMC7140484
- DOI: 10.3390/cells9030540
Translational Regulations in Response to Endoplasmic Reticulum Stress in Cancers
Abstract
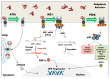
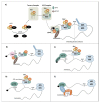
During carcinogenesis, almost all the biological processes are modified in one way or another. Among these biological processes affected, anomalies in protein synthesis are common in cancers. Indeed, cancer cells are subjected to a wide range of stresses, which include physical injuries, hypoxia, nutrient starvation, as well as mitotic, oxidative or genotoxic stresses. All of these stresses will cause the accumulation of unfolded proteins in the Endoplasmic Reticulum (ER), which is a major organelle that is involved in protein synthesis, preservation of cellular homeostasis, and adaptation to unfavourable environment. The accumulation of unfolded proteins in the endoplasmic reticulum causes stress triggering an unfolded protein response in order to promote cell survival or to induce apoptosis in case of chronic stress. Transcription and also translational reprogramming are tightly controlled during the unfolded protein response to ensure selective gene expression. The majority of stresses, including ER stress, induce firstly a decrease in global protein synthesis accompanied by the induction of alternative mechanisms for initiating the translation of mRNA, later followed by a translational recovery. After a presentation of ER stress and the UPR response, we will briefly present the different modes of translation initiation, then address the specific translational regulatory mechanisms acting during reticulum stress in cancers and highlight the importance of translational control by ER stress in tumours.
Keywords: ER stress; translation initiation; uORF; unfolded protein response (UPR), IRES.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

