Biofilm Interactions of Candida albicans and Mitis Group Streptococci in a Titanium-Mucosal Interface Model
- PMID: 32111586
- PMCID: PMC7170471
- DOI: 10.1128/AEM.02950-19
Biofilm Interactions of Candida albicans and Mitis Group Streptococci in a Titanium-Mucosal Interface Model
Abstract
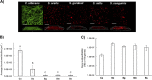
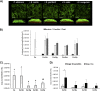
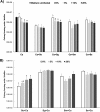
Streptococci from the mitis group (represented mainly by Streptococcus mitis, Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii) form robust biofilms with Candida albicans in different experimental models. These microorganisms have been found in polymicrobial biofilms forming on titanium biomaterial surfaces in humans with peri-implant disease. The purpose of this work was to study mutualistic interactions in biofilms forming on titanium and their effect on the adjacent mucosa, using a relevant infection model. Single and mixed biofilms of C. albicans and each Streptococcus species were grown on titanium disks. Bacterial and fungal biovolume and biomass were quantified in these biofilms. Organotypic mucosal constructs were exposed to preformed titanium surface biofilms to test their effect on secretion of proinflammatory cytokines and cell damage. C. albicans promoted bacterial biofilms of all mitis Streptococcus species on titanium surfaces. This relationship was mutualistic since all bacterial species upregulated the efg1 hypha-associated gene in C. albicans Mixed biofilms caused increased tissue damage but did not increase proinflammatory cytokine responses compared to biofilms comprising Candida alone. Interestingly, spent culture medium from tissues exposed to titanium biofilms suppressed Candida growth on titanium surfaces.IMPORTANCE Our findings provide new insights into the cross-kingdom interaction between C. albicans and Streptococcus species representative of the mitis group. These microorganisms colonize titanium-based dental implant materials, but little is known about their ability to cause inflammation and damage of the adjacent mucosal tissues. Using an in vitro biomaterial-mucosal interface infection model, we showed that mixed biofilms of each species with C. albicans enhance tissue damage. One possible mechanism for this effect is the increased fungal hypha-associated virulence gene expression we observed in mixed biofilms with these species. Interestingly, we also found that the interaction of multispecies biofilms with organotypic mucosal surfaces led to the release of growth-suppressing mediators of Candida, which may represent a homeostatic defense mechanism of the oral mucosa against fungal overgrowth. Thus, our findings provide novel insights into biofilms on biomaterials that may play an important role in the pathogenesis of mucosal infections around titanium implants.
Keywords: Candida albicans; Streptococcus; biofilms; peri-implant mucositis; peri-implantitis.
Copyright © 2020 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

