SHORTROOT-Mediated Intercellular Signals Coordinate Phloem Development in Arabidopsis Roots
- PMID: 32111671
- PMCID: PMC7203941
- DOI: 10.1105/tpc.19.00455
SHORTROOT-Mediated Intercellular Signals Coordinate Phloem Development in Arabidopsis Roots
Abstract
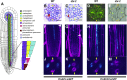
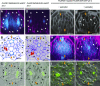
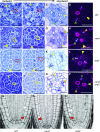
Asymmetric cell division (ACD) and positional signals play critical roles in the tissue patterning process. In the Arabidopsis (Arabidopsis thaliana) root meristem, two major phloem cell types arise via ACDs of distinct origins: one for companion cells (CCs) and the other for proto- and metaphloem sieve elements (SEs). The molecular mechanisms underlying each of these processes have been reported; however, how these are coordinated has remained elusive. Here, we report a new phloem development process coordinated via the SHORTROOT (SHR) transcription factor in Arabidopsis. The movement of SHR into the endodermis regulates the ACD for CC formation by activating microRNA165/6, while SHR moving into the phloem regulates the ACD generating the two phloem SEs. In the phloem, SHR sequentially activates NAC-REGULATED SEED MORPHOLOGY 1 (NARS1) and SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 2 (SND2), and these three together form a positive feedforward loop. Under this regulatory scheme, NARS1, generated in the CCs of the root differentiation zone, establishes a top-down signal that drives the ACD for phloem SEs in the meristem. SND2 appears to function downstream to amplify NARS1 via positive feedback. This new regulatory mechanism expands our understanding of the sophisticated vascular tissue patterning processes occurring during postembryonic root development.plantcell;32/5/1519/FX1F1fx1.
© 2020 American Society of Plant Biologists. All rights reserved.
Figures






Comment in
-
It's All in the Neighborhood: SHORTROOT-Mediated Intercellular Signals Coordinate Phloem Development in the Root.Plant Cell. 2020 May;32(5):1350-1351. doi: 10.1105/tpc.20.00156. Epub 2020 Mar 4. Plant Cell. 2020. PMID: 32132130 Free PMC article. No abstract available.
References
-
- Abrash E.B., Bergmann D.C.(2009). Asymmetric cell divisions: a view from plant development. Dev. Cell 16: 783–796. - PubMed
-
- Baum S.F., Dubrovsky J.G., Rost T.L.(2002). Apical organization and maturation of the cortex and vascular cylinder in Arabidopsis thaliana (Brassicaceae) roots. Am. J. Bot. 89: 908–920. - PubMed
-
- Berger F., Haseloff J., Schiefelbein J., Dolan L.(1998). Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Curr. Biol. 8: 421–430. - PubMed
-
- Birnbaum K., Jung J.W., Wang J.Y., Lambert G.M., Hirst J.A., Galbraith D.W., Benfey P.N.(2005). Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2: 615–619. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

