The tetraspanin Tspan15 is an essential subunit of an ADAM10 scissor complex
- PMID: 32111735
- PMCID: PMC7476718
- DOI: 10.1074/jbc.RA120.012601
The tetraspanin Tspan15 is an essential subunit of an ADAM10 scissor complex
Abstract
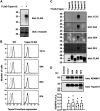
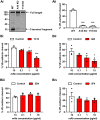
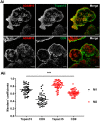
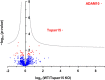
A disintegrin and metalloprotease 10 (ADAM10) is a transmembrane protein essential for embryonic development, and its dysregulation underlies disorders such as cancer, Alzheimer's disease, and inflammation. ADAM10 is a "molecular scissor" that proteolytically cleaves the extracellular region from >100 substrates, including Notch, amyloid precursor protein, cadherins, growth factors, and chemokines. ADAM10 has been recently proposed to function as six distinct scissors with different substrates, depending on its association with one of six regulatory tetraspanins, termed TspanC8s. However, it remains unclear to what degree ADAM10 function critically depends on a TspanC8 partner, and a lack of monoclonal antibodies specific for most TspanC8s has hindered investigation of this question. To address this knowledge gap, here we designed an immunogen to generate the first monoclonal antibodies targeting Tspan15, a model TspanC8. The immunogen was created in an ADAM10-knockout mouse cell line stably overexpressing human Tspan15, because we hypothesized that expression in this cell line would expose epitopes that are normally blocked by ADAM10. Following immunization of mice, this immunogen strategy generated four Tspan15 antibodies. Using these antibodies, we show that endogenous Tspan15 and ADAM10 co-localize on the cell surface, that ADAM10 is the principal Tspan15-interacting protein, that endogenous Tspan15 expression requires ADAM10 in cell lines and primary cells, and that a synthetic ADAM10/Tspan15 fusion protein is a functional scissor. Furthermore, two of the four antibodies impaired ADAM10/Tspan15 activity. These findings suggest that Tspan15 directly interacts with ADAM10 in a functional scissor complex.
Keywords: ADAM; ADAM10; Tspan14; Tspan15; a disintegrin and metalloprotease; membrane protein; metalloproteinase; molecular cell biology; monoclonal antibody; shedding; tetraspanin.
© 2020 Koo et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

