Insight into DNA substrate specificity of PARP1-catalysed DNA poly(ADP-ribosyl)ation
- PMID: 32111879
- PMCID: PMC7048826
- DOI: 10.1038/s41598-020-60631-0
Insight into DNA substrate specificity of PARP1-catalysed DNA poly(ADP-ribosyl)ation
Abstract
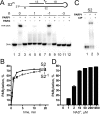
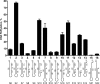
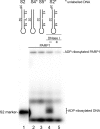
DNA-dependent poly(ADP-ribose) polymerases (PARPs) PARP1, PARP2 and PARP3 act as DNA break sensors signalling DNA damage. Upon detecting DNA damage, these PARPs use nicotine adenine dinucleotide as a substrate to synthesise a monomer or polymer of ADP-ribose (MAR or PAR, respectively) covalently attached to the acceptor residue of target proteins. Recently, it was demonstrated that PARP1-3 proteins can directly ADP-ribosylate DNA breaks by attaching MAR and PAR moieties to terminal phosphates. Nevertheless, little is still known about the mechanisms governing substrate recognition and specificity of PARP1, which accounts for most of cellular PARylation activity. Here, we characterised PARP1-mediated DNA PARylation of DNA duplexes containing various types of breaks at different positions. The 3'-terminal phosphate residue at double-strand DNA break ends served as a major acceptor site for PARP1-catalysed PARylation depending on the orientation and distance between DNA strand breaks in a single DNA molecule. A preference for ADP-ribosylation of DNA molecules containing 3'-terminal phosphate over PARP1 auto-ADP-ribosylation was observed, and a model of DNA modification by PARP1 was proposed. Similar results were obtained with purified recombinant PARP1 and HeLa cell-free extracts. Thus, the biological effects of PARP-mediated ADP-ribosylation may strongly depend on the configuration of complex DNA strand breaks.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

