Long noncoding RNA HCP5 participates in premature ovarian insufficiency by transcriptionally regulating MSH5 and DNA damage repair via YB1
- PMID: 32112110
- PMCID: PMC7192606
- DOI: 10.1093/nar/gkaa127
Long noncoding RNA HCP5 participates in premature ovarian insufficiency by transcriptionally regulating MSH5 and DNA damage repair via YB1
Abstract
The genetic etiology of premature ovarian insufficiency (POI) has been well established to date, however, the role of long noncoding RNAs (lncRNAs) in POI is largely unknown. In this study, we identified a down-expressed lncRNA HCP5 in granulosa cells (GCs) from biochemical POI (bPOI) patients, which impaired DNA damage repair and promoted apoptosis of GCs. Mechanistically, we discovered that HCP5 stabilized the interaction between YB1 and its partner ILF2, which could mediate YB1 transferring into the nucleus of GCs. HCP5 silencing affected the localization of YB1 into nucleus and reduced the binding of YB1 to the promoter of MSH5 gene, thereby diminishing MSH5 expression. Taken together, we identified that the decreased expression of HCP5 in bPOI contributed to dysfunctional GCs by regulating MSH5 transcription and DNA damage repair via the interaction with YB1, providing a novel epigenetic mechanism for POI pathogenesis.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
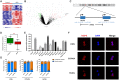

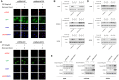

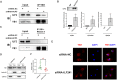

References
-
- Coulam C.B., Adamson S.C., Annegers J.F.. Incidence of premature ovarian failure. Obstet. Gynecol. 1986; 67:604–606. - PubMed
-
- Welt C.K. Primary ovarian insufficiency: a more accurate term for premature ovarian failure. Clin. Endocrinol. (Oxf.). 2008; 68:499–509. - PubMed
-
- Jiao X., Ke H., Qin Y., Chen Z.J.. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol. Metab. 2018; 29:795–807. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

