Characterization of 100 extended major histocompatibility complex haplotypes in Indonesian cynomolgus macaques
- PMID: 32112172
- PMCID: PMC7223175
- DOI: 10.1007/s00251-020-01159-5
Characterization of 100 extended major histocompatibility complex haplotypes in Indonesian cynomolgus macaques
Abstract
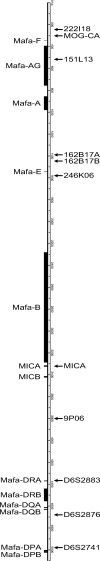
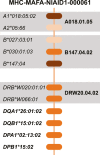
Many medical advancements-including improvements to anti-rejection therapies in transplantation and vaccine development-rely on preclinical studies conducted in cynomolgus macaques (Macaca fascicularis). Major histocompatibility complex (MHC) class I and class II genes of cynomolgus macaques are orthologous to human leukocyte antigen complex (HLA) class I and class II genes, respectively. Both encode cell-surface proteins involved in cell recognition and rejection of non-host tissues. MHC class I and class II genes are highly polymorphic, so comprehensive genotyping requires the development of complete databases of allelic variants. Our group used PacBio circular consensus sequencing of full-length cDNA amplicons to characterize MHC class I and class II transcript sequences for a cohort of 293 Indonesian cynomolgus macaques (ICM) in a large, pedigreed breeding colony. These studies allowed us to expand the existing database of Macaca fascicularis (Mafa) alleles by identifying an additional 141 MHC class I and 61 class II transcript sequences. In addition, we defined co-segregating combinations of allelic variants as regional haplotypes for 70 Mafa-A, 78 Mafa-B, and 45 Mafa-DRB gene clusters. Finally, we defined class I and class II transcripts that are associated with 100 extended MHC haplotypes in this breeding colony by combining our genotyping analyses with short tandem repeat (STR) patterns across the MHC region. Our sequencing analyses and haplotype definitions improve the utility of these ICM for transplantation studies as well as infectious disease and vaccine research.
Keywords: Haplotype; Macaca fascicularis; Major histocompatibility complex; Short tandem repeat; Transplantation.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures


References
-
- Aarnink A, Mee ET, Savy N, Congy-Jolivet N, Rose NJ, Blancher A. Deleterious impact of feto-maternal MHC compatibility on the success of pregnancy in a macaque model. Immunogenetics. 2014;66(2):105–113. - PubMed
-
- Bruijnesteijn J, van der Wiel MKH, Swelsen WTN, Otting N, de Vos-Rouweler AJM, Elferink D, Doxiadis GG, Claas FHJ, Lardy NM, de Groot NG, Bontrop RE. Human and rhesus macaque KIR haplotypes defined by their transcriptomes. J Immunol. 2018;200(5):1692–1701. - PubMed
Publication types
MeSH terms
Grants and funding
- P51OD011106/National Institutes of Health Office of Research Infrastructure Programs/International
- RR020141-01/Foundation for the National Institutes of Health/International
- RR15459-01/Foundation for the National Institutes of Health/International
- T32 GM008692/GM/NIGMS NIH HHS/United States
- T15 LM007359/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

