Enhancing reaction-based de novo design using a multi-label reaction class recommender
- PMID: 32112286
- PMCID: PMC7293200
- DOI: 10.1007/s10822-020-00300-6
Enhancing reaction-based de novo design using a multi-label reaction class recommender
Abstract
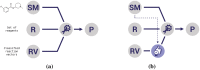
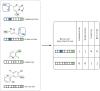
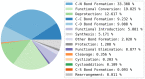
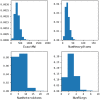
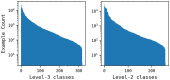
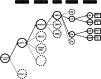
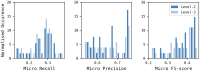
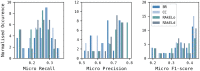
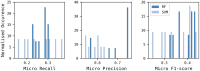
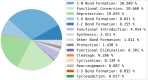
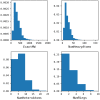
Reaction-based de novo design refers to the in-silico generation of novel chemical structures by combining reagents using structural transformations derived from known reactions. The driver for using reaction-based transformations is to increase the likelihood of the designed molecules being synthetically accessible. We have previously described a reaction-based de novo design method based on reaction vectors which are transformation rules that are encoded automatically from reaction databases. A limitation of reaction vectors is that they account for structural changes that occur at the core of a reaction only, and they do not consider the presence of competing functionalities that can compromise the reaction outcome. Here, we present the development of a Reaction Class Recommender to enhance the reaction vector framework. The recommender is intended to be used as a filter on the reaction vectors that are applied during de novo design to reduce the combinatorial explosion of in-silico molecules produced while limiting the generated structures to those which are most likely to be synthesisable. The recommender has been validated using an external data set extracted from the recent medicinal chemistry literature and in two simulated de novo design experiments. Results suggest that the use of the recommender drastically reduces the number of solutions explored by the algorithm while preserving the chance of finding relevant solutions and increasing the global synthetic accessibility of the designed molecules.
Keywords: De novo design; Multi-label classification; Reaction class recommender; Reaction vector.
Figures
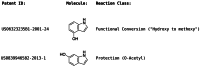
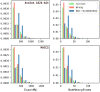

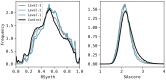
References
-
- Hartenfeller M, Schneider G, Hartenfeller M, Proschak E. De novo drug design. In: Bajorath J, editor. Lead generation approaches in drug discovery. Hoboken: Wiley; 2010. pp. 165–185.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

