Imaging of intratumoral heterogeneity in high-grade glioma
- PMID: 32112907
- PMCID: PMC7108976
- DOI: 10.1016/j.canlet.2020.02.025
Imaging of intratumoral heterogeneity in high-grade glioma
Abstract
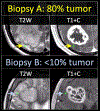
High-grade glioma (HGG), and particularly Glioblastoma (GBM), can exhibit pronounced intratumoral heterogeneity that confounds clinical diagnosis and management. While conventional contrast-enhanced MRI lacks the capability to resolve this heterogeneity, advanced MRI techniques and PET imaging offer a spectrum of physiologic and biophysical image features to improve the specificity of imaging diagnoses. Published studies have shown how integrating these advanced techniques can help better define histologically distinct targets for surgical and radiation treatment planning, and help evaluate the regional heterogeneity of tumor recurrence and response assessment following standard adjuvant therapy. Application of texture analysis and machine learning (ML) algorithms has also enabled the emerging field of radiogenomics, which can spatially resolve the regional and genetically distinct subpopulations that coexist within a single GBM tumor. This review focuses on the latest advances in neuro-oncologic imaging and their clinical applications for the assessment of intratumoral heterogeneity.
Keywords: Advanced; Glioblastoma; Glioma; Heterogeneity; Histologic; Imaging; Intratumoral; MRI; Radiogenomics.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest US Patents: US8571844B2 (KRS). US Patent Applications: 15/290,963 (LSH, JL); PCT/US2018/061887 (LSH, AHD, JL, KRS); PCT/US2019/019687 (LSH, JL, KRS). Grant funding: Funding: NS082609 (NIH), CA221938 (NIH), CA220378 (NIH), Mayo Clinic Foundation (U.S.A.), James S. McDonnell Foundation (U.S.A), Ben and Catherine Ivy Foundation (U.S.A.), Arizona Biomedical Research Commission (U.S.A.).
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

