Role of the DNA repair genes H2AX and HMGB1 in human fat distribution and lipid profiles
- PMID: 32114485
- PMCID: PMC7050360
- DOI: 10.1136/bmjdrc-2019-000831
Role of the DNA repair genes H2AX and HMGB1 in human fat distribution and lipid profiles
Abstract
Introduction: Regional fat distribution strongly relates to metabolic comorbidities. We identified the DNA repair genes H2AX and HMGB1 to be differentially expressed between human subcutaneous (SAT) and omental visceral adipose tissue (OVAT) depots. As increased DNA damage is linked to metabolic disease, we here sought to analyze whether depot-specific H2AX and HMGB1 expression is related to anthropometric and metabolic profiles of obesity. We further tested for different H2AX mRNA regulatory mechanisms by analyzing promoter DNA methylation and genotyped rs7350 in the H2AX locus.
Research design and methods: Gene expression (OVAT n=48; SAT n=55) and DNA promoter methylation data (OVAT and SAT n=77) were extracted from an existing dataset as described elsewhere. Genotype data for the 3'untranslated region (3'UTR) H2AX variant rs7350 were generated by using the TaqMan genotyping system in 243 subjects of the same cohort. Statistical analyses were done using SPSS statistics software 24 and GraphPad Prism 6.
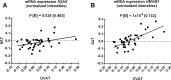
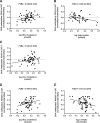
Results: We identified H2AX being higher (p=0.002) and HMGB1 being less expressed (p=0.0001) in OVAT compared with SAT. Further, we observed positive interdepot correlations of OVAT and SAT for both HMGB1 (p=1×10-6) and H2AX mRNA levels (p=0.024). Depot-specific associations were observed for both genes' methylation levels with either high density lipoprotein cholesterol, low density lipoprotein cholesterol, triglycerides and/or with OVAT/SAT-ratio (all p<0.05). A significantly lower level of total cholesterol in minor A-Allele carriers of rs7350 compared with AG and GG carriers (p=0.001) was observed. Additionally, subjects carrying the A-allele showed lower SAT HMGB1 expression level (p=0.030).
Conclusion: Our results suggest a fat depot-specific regulation of H2AX and HMGB1 potentially mediated by both DNA methylation and genetic variation. Rs7350, DNA methylation and/or mRNA levels of H2AX and HMGB1 are related to lipid parameters. Further studies are warranted to evaluate the functional role of the DNA repair genes H2AX and HMGB1 in obesity and fat distribution.
Keywords: fat distribution; metabolic syndrome; obesity and body fat distribution; transcription.
© Author(s) (or their employer(s)) 2020. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
