Transcriptome profiling of Ewing sarcomas - treatment resistance pathways and IGF-dependency
- PMID: 32115849
- PMCID: PMC7191197
- DOI: 10.1002/1878-0261.12655
Transcriptome profiling of Ewing sarcomas - treatment resistance pathways and IGF-dependency
Abstract
Ewing sarcomas (ESs) are aggressive sarcomas driven by EWS fusion genes. We sought to investigate whether whole-transcriptome sequencing (RNA-seq) could be used to detect patterns associated with chemotherapy response or tumor progression after first-line treatment. Transcriptome sequencing (RNA-seq) of 13 ES cases was performed. Among the differentially expressed pathways, we identified IGF2 expression as a potential driver of chemotherapy response and progression. We investigated the effect of IGF2 on proliferation, radioresistance, apoptosis, and the transcriptome pattern in four ES cell lines and the effect of IGF2 expression in a validation series of 14 patients. Transcriptome analysis identified differentially expressed genes (adj. P < 0.005) and pathways associated with chemotherapy response (285 genes), short overall survival (662 genes), and progression after treatment (447 genes). Imprinting independent promoter P3-mediated IGF2 expression was identified in a subset of cases with aggressive clinical course. In ES cell lines, IGF2 induced proliferation, but promoted radioresistance only in CADO cells. High IGF2 expression was also significantly associated with shorter overall survival in patients with ES. Transcriptome analysis of the clinical samples and the cell lines revealed an IGF-dependent signature, potentially related to a stem cell-like phenotype. Transcriptome analysis is a potentially powerful complementary tool to predict the clinical behavior of ES and may be utilized for clinical trial stratification strategies and personalized oncology. Certain gene signatures, for example, IGF-related pathways, are coupled to biological functions that could be of clinical importance. Finally, our results indicate that IGF inhibition may be successful as a first-line therapy in conjunction with conventional radiochemotherapy for a subset of patients.
Keywords: Ewing sarcoma; IGF2; RNA-seq; apoptosis; transcriptome profiling; tumor progression.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
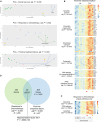

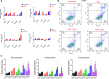


References
-
- Crompton BD, Stewart C, Taylor‐Weiner A, Alexe G, Kurek KC, Calicchio ML, Kiezun A, Carter SL, Shukla SA, Mehta SS et al (2014) The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 4, 1326–1341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

