Salinity tolerance in barley during germination- homologs and potential genes
- PMID: 32115909
- PMCID: PMC7076347
- DOI: 10.1631/jzus.B1900400
Salinity tolerance in barley during germination- homologs and potential genes
Abstract
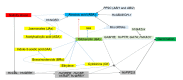
Salinity affects more than 6% of the world's total land area, causing massive losses in crop yield. Salinity inhibits plant growth and development through osmotic and ionic stresses; however, some plants exhibit adaptations through osmotic regulation, exclusion, and translocation of accumulated Na+ or Cl-. Currently, there are no practical, economically viable methods for managing salinity, so the best practice is to grow crops with improved tolerance. Germination is the stage in a plant's life cycle most adversely affected by salinity. Barley, the fourth most important cereal crop in the world, has outstanding salinity tolerance, relative to other cereal crops. Here, we review the genetics of salinity tolerance in barley during germination by summarizing reported quantitative trait loci (QTLs) and functional genes. The homologs of candidate genes for salinity tolerance in Arabidopsis, soybean, maize, wheat, and rice have been blasted and mapped on the barley reference genome. The genetic diversity of three reported functional gene families for salt tolerance during barley germination, namely dehydration-responsive element-binding (DREB) protein, somatic embryogenesis receptor-like kinase and aquaporin genes, is discussed. While all three gene families show great diversity in most plant species, the DREB gene family is more diverse in barley than in wheat and rice. Further to this review, a convenient method for screening for salinity tolerance at germination is needed, and the mechanisms of action of the genes involved in salt tolerance need to be identified, validated, and transferred to commercial cultivars for field production in saline soil.
Keywords: Genetics; Barley; Quantitative trait locus (QTL); Germination; Salinity tolerance; Homologous gene; Diversity.
Conflict of interest statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Figures
Similar articles
-
Genome-Wide Association Study of Salt Tolerance-Related Traits during Germination and Seedling Development in an Intermedium-Spike Barley Collection.Int J Mol Sci. 2022 Sep 21;23(19):11060. doi: 10.3390/ijms231911060. Int J Mol Sci. 2022. PMID: 36232362 Free PMC article.
-
Dissecting new genetic components of salinity tolerance in two-row spring barley at the vegetative and reproductive stages.PLoS One. 2020 Jul 23;15(7):e0236037. doi: 10.1371/journal.pone.0236037. eCollection 2020. PLoS One. 2020. PMID: 32701981 Free PMC article.
-
Genome-Wide Association Study of Salinity Tolerance During Germination in Barley (Hordeum vulgare L.).Front Plant Sci. 2020 Feb 21;11:118. doi: 10.3389/fpls.2020.00118. eCollection 2020. Front Plant Sci. 2020. PMID: 32153619 Free PMC article.
-
Drought and salt tolerances in wild relatives for wheat and barley improvement.Plant Cell Environ. 2010 Apr;33(4):670-85. doi: 10.1111/j.1365-3040.2009.02107.x. Epub 2010 Feb 5. Plant Cell Environ. 2010. PMID: 20040064 Review.
-
Salinity stress tolerance and omics approaches: revisiting the progress and achievements in major cereal crops.Heredity (Edinb). 2022 Jun;128(6):497-518. doi: 10.1038/s41437-022-00516-2. Epub 2022 Mar 5. Heredity (Edinb). 2022. PMID: 35249098 Free PMC article. Review.
Cited by
-
Red light enhances folate accumulation in wheat seedlings.J Zhejiang Univ Sci B. 2021 Nov 15;22(11):906-916. doi: 10.1631/jzus.B2100266. J Zhejiang Univ Sci B. 2021. PMID: 34783221 Free PMC article. English.
-
The utilization of microbes for sustainable food production.BioTechnologia (Pozn). 2023 Jun 26;104(2):209-216. doi: 10.5114/bta.2023.127209. eCollection 2023. BioTechnologia (Pozn). 2023. PMID: 37427028 Free PMC article. Review.
-
Applications of cell- and tissue-specific 'omics to improve plant productivity.Emerg Top Life Sci. 2022 Apr 15;6(2):163-173. doi: 10.1042/ETLS20210286. Emerg Top Life Sci. 2022. PMID: 35293572 Free PMC article. Review.
-
Breeding crops by design for future agriculture.J Zhejiang Univ Sci B. 2020 Jun;21(6):423-425. doi: 10.1631/jzus.B2010001. J Zhejiang Univ Sci B. 2020. PMID: 32478489 Free PMC article.
-
Germination and Growth Characteristics of nud Knockout and win1 Knockout Barley Lines under Salt Stress.Plants (Basel). 2024 Apr 23;13(9):1169. doi: 10.3390/plants13091169. Plants (Basel). 2024. PMID: 38732384 Free PMC article.
References
-
- Abido WAE, Allem A, Zsombic L, et al. Effect of gibberellic acid on germination of six wheat cultivars under salinity stress levels. Asian J Biol Sci. 2019;12(1):51–60. doi: 10.3923/ajbs.2019.51.60. - DOI
-
- Abrol IP, Yadav JSP, Massoud FI. Salt-Affected Soils and Their Management. FAO Soils Bulletin 39, Food and Agriculture Organization of the United Nations, Rome; 1988.
-
- Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol Plant. 2010;54(2):201–212. doi: 10.1007/s10535-010-0038-7. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

