Characterizing Adult Cochlear Supporting Cell Transcriptional Diversity Using Single-Cell RNA-Seq: Validation in the Adult Mouse and Translational Implications for the Adult Human Cochlea
- PMID: 32116546
- PMCID: PMC7012811
- DOI: 10.3389/fnmol.2020.00013
Characterizing Adult Cochlear Supporting Cell Transcriptional Diversity Using Single-Cell RNA-Seq: Validation in the Adult Mouse and Translational Implications for the Adult Human Cochlea
Erratum in
-
Corrigendum: Characterizing Adult Cochlear Supporting Cell Transcriptional Diversity Using Single-Cell RNA-Seq: Validation in the Adult Mouse and Translational Implications for the Adult Human Cochlea.Front Mol Neurosci. 2021 Jun 3;14:699859. doi: 10.3389/fnmol.2021.699859. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34149357 Free PMC article.
Abstract
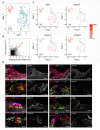
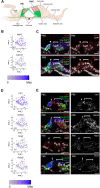
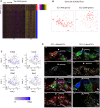
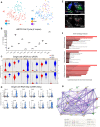
Hearing loss is a problem that impacts a significant proportion of the adult population. Cochlear hair cell (HC) loss due to loud noise, chemotherapy and aging is the major underlying cause. A significant proportion of these individuals are dissatisfied with available treatment options which include hearing aids and cochlear implants. An alternative approach to restore hearing would be to regenerate HCs. Such therapy would require a recapitulation of the complex architecture of the organ of Corti, necessitating regeneration of both mature HCs and supporting cells (SCs). Transcriptional profiles of the mature cell types in the cochlea are necessary to can provide a metric for eventual regeneration therapies. To assist in this effort, we sought to provide the first single-cell characterization of the adult cochlear SC transcriptome. We performed single-cell RNA-Seq on FACS-purified adult cochlear SCs from the LfngEGFP adult mouse, in which SCs express GFP. We demonstrate that adult cochlear SCs are transcriptionally distinct from their perinatal counterparts. We establish cell-type-specific adult cochlear SC transcriptome profiles, and we validate these expression profiles through a combination of both fluorescent immunohistochemistry and in situ hybridization co-localization and quantitative polymerase chain reaction (qPCR) of adult cochlear SCs. Furthermore, we demonstrate the relevance of these profiles to the adult human cochlea through immunofluorescent human temporal bone histopathology. Finally, we demonstrate cell cycle regulator expression in adult SCs and perform pathway analyses to identify potential mechanisms for facilitating mitotic regeneration (cell proliferation, differentiation, and eventually regeneration) in the adult mammalian cochlea. Our findings demonstrate the importance of characterizing mature as opposed to perinatal SCs.
Keywords: FACS; adult (MeSH); cell cycle; cochlea; inner ear; smFISH; supporting cell subtypes.
Copyright © 2020 Hoa, Olszewski, Li, Taukulis, Gu, DeTorres, Lopez, Linthicum, Ishiyama, Martin, Morell and Kelley.
Figures

References
-
- Blondel V. D., Guillaume J.-L., Lambiotte R., Lefebvre E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008:P10008. 10.1088/1742-5468/2008/10/P10008 - DOI
-
- Chen P., Segil N. (1999). P27(Kip1) links cell proliferation to morphogenesis in the developing organ of corti. Development 126, 1581–1590. - PubMed

