Clonal Selection of a Novel Deleterious TP53 Somatic Mutation Discovered in ctDNA of a KIT/PDGFRA Wild-Type Gastrointestinal Stromal Tumor Resistant to Imatinib
- PMID: 32116712
- PMCID: PMC7019050
- DOI: 10.3389/fphar.2020.00036
Clonal Selection of a Novel Deleterious TP53 Somatic Mutation Discovered in ctDNA of a KIT/PDGFRA Wild-Type Gastrointestinal Stromal Tumor Resistant to Imatinib
Abstract
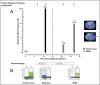
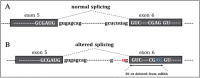
The standard of care for the first-line treatment of advanced gastrointestinal stromal tumor (GIST) is represented by imatinib, which is given daily at a standard dosage until tumor progression. Resistance to imatinib commonly occurs through the clonal selection of genetic mutations in the tumor DNA, and an increase in imatinib dosage was demonstrated to be efficacious to overcome imatinib resistance. Wild-type GISTs, which do not display KIT or platelet-derived growth factor receptor alpha (PDGFRA) mutations, are usually primarily insensitive to imatinib and tend to rapidly relapse in course of treatment. Here we report the case of a 53-year-old male patient with gastric GIST who primarily did not respond to imatinib and that, despite the administration of an increased imatinib dose, led to patient death. By using a deep next-generation sequencing barcode-aware approach, we analyzed a panel of actionable cancer-related genes in the patient cfDNA to investigate somatic changes responsible for imatinib resistance. We identified, in two serial circulating tumor DNA (ctDNA) samples, a sharp increase in the allele frequency of a never described TP53 mutation (c.560-7_560-2delCTCTTAinsT) located in a splice acceptor site and responsible for a protein loss of function. The same TP53 mutation was retrospectively identified in the primary tumor by digital droplet PCR at a subclonal frequency (0.1%). The mutation was detected at a very high allelic frequency (99%) in the metastatic hepatic lesion, suggesting a rapid clonal selection of the mutation during tumor progression. Imatinib plasma concentration at steady state was above the threshold of 760 ng/ml reported in the literature for the minimum efficacious concentration. The de novo TP53 (c.560-7_560-2delCTCTTAinsT) mutation was in silico predicted to be associated with an aberrant RNA splicing and with an aggressive phenotype which might have contributed to a rapid disease spread despite the administration of an increased imatinib dosage. This result underlies the need of a better investigation upon the role of TP53 in the pathogenesis of GISTs and sustains the use of next-generation sequencing (NGS) in cfDNA for the identification of novel genetic markers in wild-type GISTs.
Keywords: TP53; circulating tumor DNA; gastrointestinal stromal tumor; imatinib; liquid biopsy.
Copyright © 2020 Dalle Fratte, Guardascione, De Mattia, Borsatti, Boschetto, Farruggio, Canzonieri, Romanato, Borsatti, Gagno, Marangon, Polano, Buonadonna, Toffoli and Cecchin.
Figures

References
-
- Bouchet S., Poulette S., Titier K., Moore N., Lassalle R., Abouelfath A., et al. (2016). Relationship between imatinib trough concentration and outcomes in the treatment of advanced gastrointestinal stromal tumours in a real-life setting. Eur. J. Cancer 57, 31–38. 10.1016/j.ejca.2015.12.029 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

