Characterization of Acinetobacter baumannii Copper Resistance Reveals a Role in Virulence
- PMID: 32117089
- PMCID: PMC7015863
- DOI: 10.3389/fmicb.2020.00016
Characterization of Acinetobacter baumannii Copper Resistance Reveals a Role in Virulence
Abstract
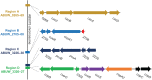
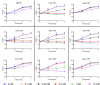
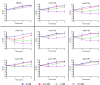
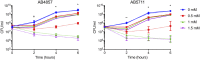
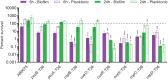
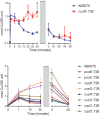
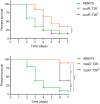
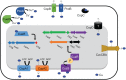
Acinetobacter baumannii is often highly drug-resistant and causes severe infections in compromised patients. These infections can be life threatening due to limited treatment options. Copper is inherently antimicrobial and increasing evidence indicates that copper containing formulations may serve as non-traditional therapeutics against multidrug-resistant bacteria. We previously reported that A. baumannii is sensitive to high concentrations of copper. To understand A. baumannii copper resistance at the molecular level, herein we identified putative copper resistance components and characterized 21 strains bearing mutations in these genes. Eight of the strains displayed a copper sensitive phenotype (pcoA, pcoB, copB, copA/cueO, copR/cusR, copS/cusS, copC, copD); the putative functions of these proteins include copper transport, oxidation, sequestration, and regulation. Importantly, many of these mutant strains still showed increased sensitivity to copper while in a biofilm. Inductively coupled plasma mass spectrometry revealed that many of these strains had defects in copper mobilization, as the mutant strains accumulated more intracellular copper than the wild-type strain. Given the crucial antimicrobial role of copper-mediated killing employed by the immune system, virulence of these mutant strains was investigated in Galleria mellonella; many of the mutant strains were attenuated. Finally, the cusR and copD strains were also investigated in the murine pneumonia model; both were found to be important for full virulence. Thus, copper possesses antimicrobial activity against multidrug-resistant A. baumannii, and copper sensitivity is further increased when copper homeostasis mechanisms are interrupted. Importantly, these proteins are crucial for full virulence of A. baumannii and may represent novel drug targets.
Keywords: Acinetobacter baumannii; Galleria; copper; metal; pathogenesis.
Copyright © 2020 Williams, Neu, Alamneh, Reddinger, Jacobs, Singh, Abu-Taleb, Michel, Zurawski and Merrell.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

