Resistome in Lake Bolonha, Brazilian Amazon: Identification of Genes Related to Resistance to Broad-Spectrum Antibiotics
- PMID: 32117110
- PMCID: PMC7010645
- DOI: 10.3389/fmicb.2020.00067
Resistome in Lake Bolonha, Brazilian Amazon: Identification of Genes Related to Resistance to Broad-Spectrum Antibiotics
Abstract
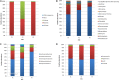
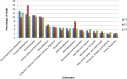
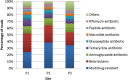
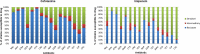
Resistance to antibiotics is one of the most relevant public health concerns in the world. Aquatic environments play an important role because they are reservoirs for antibiotic resistance genes and antibiotic-resistant strains, contributing to the spread of resistance. The present study investigated the resistome in Lake Bolonha (three sampling sites) in the Amazon region using a metagenomics approach and culture-dependent methods. Whole-metagenome-based results showed that the most abundant phyla were Protobacteria, Actinobacteria, Firmicutes, Bacteroidetes and Cyanobacteria. The composition of the resistome demonstrated that the genes that confer resistance to β-lactams were prevalent at all sampling sites, followed by genes conferring resistance to aminoglycosides and tetracycline. Acquired genes encoding extended-spectrum β-lactamases (e.g., bla CTX-M) and resistance to carbapenems (e.g., bla IMP and bla VIM) were detected through metagenome analysis. Bacteria were isolated from culture medium supplemented with cefotaxime or imipenem, and isolates were identified and analyzed for their antibiotic susceptibility profiles and resistance genes. In total, 98 bacterial isolates belonging to the genera Pseudomonas (37), Acinetobacter (32), Klebsiella (13), Enterobacter (9), Pantoe (3), Stenotrophomonas (3), and Methylobacterium (1) were obtained. Among isolates, the most abundant genes were bla CTX-M (28.3%), bla SHV (22.6%) and bla TEM (18.8%) in isolates from cefotaxime-supplemented medium and bla VIM (28.8%) and bla IMP (22.2%) in isolates recovered from imipenem-supplemented medium. The genes intl1 and intl2 were detected in 19.3% and 7.1% of isolates. Antibiograms showed that 94.9% (from cefotaxime-supplemented medium) and 85.7% (from imipenem-supplemented medium) of the isolates were multidrug resistant. Besides cefotaxime and imipenem, isolates were mostly resistant to aztreonam (91.8%), amoxicillin (98.8%), ampicillin (82.6%), and nalidixic acid (77.5%). Hence, the present study demonstrates that Lake Bolonha is a reservoir of bacteria resistant to antibiotics and resistance genes, some of which are of critical importance to human health.
Keywords: Lake Bolonha; bacteria resistant to antibiotics; metagenome; resistome; β-lactams.
Copyright © 2020 Alves, Dias, Mateus, Marques, Graças, Ramos, Seldin, Henriques, Silva and Folador.
Figures
Similar articles
-
Extended Spectrum Beta-Lactamase-Producing Gram-Negative Bacteria Recovered From an Amazonian Lake Near the City of Belém, Brazil.Front Microbiol. 2019 Feb 28;10:364. doi: 10.3389/fmicb.2019.00364. eCollection 2019. Front Microbiol. 2019. PMID: 30873145 Free PMC article.
-
Occurrence of IMP-8, IMP-10, and IMP-13 metallo-β-lactamases located on class 1 integrons and other extended-spectrum β-lactamases in bacterial isolates from Tunisian rivers.Scand J Infect Dis. 2013 Feb;45(2):95-103. doi: 10.3109/00365548.2012.717712. Epub 2012 Sep 19. Scand J Infect Dis. 2013. PMID: 22992193
-
Distribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran.Microb Drug Resist. 2010 Mar;16(1):49-53. doi: 10.1089/mdr.2009.0096. Microb Drug Resist. 2010. PMID: 19961397
-
Ultrastructural Changes in Clinical and Microbiota Isolates of Klebsiella pneumoniae Carriers of Genes bla SHV, bla TEM, bla CTX-M, or bla KPC When Subject to β-Lactam Antibiotics.ScientificWorldJournal. 2015;2015:572128. doi: 10.1155/2015/572128. Epub 2015 Sep 28. ScientificWorldJournal. 2015. PMID: 26491715 Free PMC article.
-
Spread of TEM, VIM, SHV, and CTX-M β-Lactamases in Imipenem-Resistant Gram-Negative Bacilli Isolated from Egyptian Hospitals.Int J Microbiol. 2016;2016:8382605. doi: 10.1155/2016/8382605. Epub 2016 Mar 31. Int J Microbiol. 2016. PMID: 27123005 Free PMC article.
Cited by
-
Genomic insights into ESBL-producing Escherichia coli isolated from non-human primates in the Peruvian Amazon.Front Vet Sci. 2024 Jan 16;10:1340428. doi: 10.3389/fvets.2023.1340428. eCollection 2023. Front Vet Sci. 2024. PMID: 38292135 Free PMC article.
-
One Health Implications of Antimicrobial Resistance in Bacteria from Amazon River Dolphins.Ecohealth. 2021 Sep;18(3):383-396. doi: 10.1007/s10393-021-01558-4. Epub 2021 Oct 28. Ecohealth. 2021. PMID: 34709509
-
Genomic insights of Klebsiella pneumoniae isolated from a native Amazonian fish reveal wide resistome against heavy metals, disinfectants, and clinically relevant antibiotics.Genomics. 2020 Nov;112(6):5143-5146. doi: 10.1016/j.ygeno.2020.09.015. Epub 2020 Sep 9. Genomics. 2020. PMID: 32916256 Free PMC article.
-
Exploring the Bacteriome and Resistome of Humans and Food-Producing Animals in Brazil.Microbiol Spectr. 2022 Oct 26;10(5):e0056522. doi: 10.1128/spectrum.00565-22. Epub 2022 Aug 22. Microbiol Spectr. 2022. PMID: 35993730 Free PMC article.
-
Distribution of Beta-Lactamase Producing Gram-Negative Bacterial Isolates in Isabela River of Santo Domingo, Dominican Republic.Front Microbiol. 2021 Jan 13;11:519169. doi: 10.3389/fmicb.2020.519169. eCollection 2020. Front Microbiol. 2021. PMID: 33519720 Free PMC article.
References
-
- Alves M. S., Pereira A., Araújo S. M., Castro B. B., Correia A. C. M., Henriques I. (2014). Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front. Microbiol. 5:426. 10.3389/fmicb.2014.00426 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

