Agricultural Selection of Wheat Has Been Shaped by Plant-Microbe Interactions
- PMID: 32117153
- PMCID: PMC7015950
- DOI: 10.3389/fmicb.2020.00132
Agricultural Selection of Wheat Has Been Shaped by Plant-Microbe Interactions
Abstract
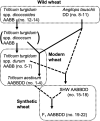
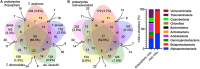
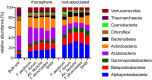
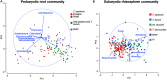
The influence of wheat (modern wheat, both bread and pasta, their wild ancestors and synthetic hybrids) on the microbiota of their roots and surrounding soil is characterized. We isolated lines of bread wheat by hybridizing diploid (Aegilops tauschii) with tetraploid Triticum durum and crossed it with a modern cultivar of Triticum aestivum. The newly created, synthetic hybrid wheat, which recapitulate the breeding history of wheat through artificial selection, is found to support a microbiome enriched in beneficial Glomeromycetes fungi, but also in, potentially detrimental, Nematoda. We hypothesize that during wheat domestication this plant-microbe interaction diminished, suggesting an evolutionary tradeoff; sacrificing advantageous nutrient acquisition through fungal interactions to minimize interaction with pathogenic fungi. Increased plant selection for Glomeromycetes and Nematoda is correlated with the D genome derived from A. tauschii. Despite differences in their soil microbiota communities, overall wheat plants consistently show a low ratio of eukaryotes to prokaryotes. We propose that this is a mechanism for protection against soil-borne fungal disease and appears to be deeply rooted in the wheat genome. We suggest that the influence of plants on the composition of their associated microbiota is an integral factor, hitherto overlooked, but intrinsic to selection during wheat domestication.
Keywords: Triticaeae; crop domestication; microbiota; polyploidy; rhizosphere; wheat.
Copyright © 2020 Tkacz, Pini, Turner, Bestion, Simmonds, Howell, Greenland, Cheema, Emms, Uauy and Poole.
Figures


References
-
- Anderson M. J., Gorley R. N., Clarke K. R. (2008). PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. Plymouth: PRIMER-E.
-
- Bergman B., Matveyev A., Rasmussen U. (1996). Chemical signalling in cyanobacterial-plant symbioses. Trends Plant Sci. 1 191–197. 10.1016/1360-1385(96)10021-2 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

