Novel Insights Into Immune Systems of Bats
- PMID: 32117225
- PMCID: PMC7025585
- DOI: 10.3389/fimmu.2020.00026
Novel Insights Into Immune Systems of Bats
Abstract
In recent years, viruses similar to those that cause serious disease in humans and other mammals have been detected in apparently healthy bats. These include filoviruses, paramyxoviruses, and coronaviruses that cause severe diseases such as Ebola virus disease, Marburg haemorrhagic fever and severe acute respiratory syndrome (SARS) in humans. The evolution of flight in bats seem to have selected for a unique set of antiviral immune responses that control virus propagation, while limiting self-damaging inflammatory responses. Here, we summarize our current understanding of antiviral immune responses in bats and discuss their ability to co-exist with emerging viruses that cause serious disease in other mammals. We highlight how this knowledge may help us to predict viral spillovers into new hosts and discuss future directions for the field.
Keywords: antiviral; bats (Chiroptera); emerging viruses; innate and adaptive immune response; interferon; virus.
Copyright © 2020 Banerjee, Baker, Kulcsar, Misra, Plowright and Mossman.
Figures
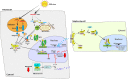
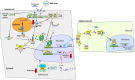
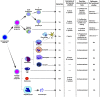
References
-
- CDC Severe Acute Respiratory Syndrome. (2004). Available online at: https://www.cdc.gov/sars/about/fs-sars.html (cited February.2.001 20, 2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

