The Immunomodulatory Properties of the Human Amnion-Derived Mesenchymal Stromal/Stem Cells Are Induced by INF-γ Produced by Activated Lymphomonocytes and Are Mediated by Cell-To-Cell Contact and Soluble Factors
- PMID: 32117234
- PMCID: PMC7028706
- DOI: 10.3389/fimmu.2020.00054
The Immunomodulatory Properties of the Human Amnion-Derived Mesenchymal Stromal/Stem Cells Are Induced by INF-γ Produced by Activated Lymphomonocytes and Are Mediated by Cell-To-Cell Contact and Soluble Factors
Abstract
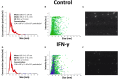
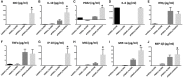
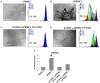
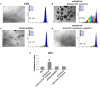
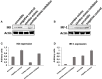
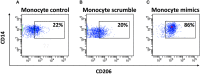
Human mesenchymal stromal/stem cells (MSCs), being immunoprivileged and having immunomodulatory ability, represent a promising tool to be applied in the field of regenerative medicine. Based on numerous in vitro evidences, the immunological effects of MSCs on immune cells could depend on different mechanisms as cell-to-cell contact and paracrine signals. Furthermore, recent studies have shown that the immunomodulatory activity of MSCs is initiated by activated immune cells; thus, their interaction represents a potential homeostatic mechanism by which MSCs regulate the immune response. MSCs also release exosomes able to give different effects, in a paracrine manner, by influencing inflammatory processes. In this study, we aimed to establish the potential role of human amnion-derived MSCs (hAMSCs), in immunomodulation. We found that the immunosuppressive properties of hAMSCs are not constitutive, but require "supportive signals" capable of promoting these properties. Indeed, we observed that hAMSCs alone are not able to produce an adequate amount of soluble immunomodulatory factors. Here, we studied, in depth, the strong immunomodulatory licensing signal deriving from the direct interaction between hAMSCs and stimulated peripheral blood mononuclear cells. We found that the immunomodulatory effect of hAMSCs also depends on cell-to-cell contact through the contribution of the PDL-1/PD-1 axis. We then investigated the IFN-γ priming of hAMSCs (γ-hAMSCs), which induce the increase of PDL-1 expression, high production of IDO, and upregulation of different immunomodulatory exosome-derived miRNAs. Our miRNA-target network analysis revealed that nine of the deregulated miRNAs are involved in the regulation of key proteins that control both T cell activation/anergy and monocyte differentiation pathways. Finally, we observed that γ-hAMSCs induce in monocytes both M2-like phenotype and the increase of IL-10 production. The extensive implications of MSCs in modulating different aspects of the immune system make these cells attractive candidates to be employed in therapeutic application in immune-based diseases. For these reasons, we aimed, with this study, to shed light on the potential of hAMSCs, and how they could become a useful tool for treating different inflammatory diseases, including end-stage pathologies or adverse effects in transplanted patients.
Keywords: M2-like monocytes; PDL-1; exosomes; human amnion-derived mesenchymal stem cells; immunomodulation; interferon-γ; primed-hAMSCs; regenerative medicine.
Copyright © 2020 Bulati, Miceli, Gallo, Amico, Carcione, Pampalone and Conaldi.
Figures





References
-
- Parolini O, Alviano F, Bagnara GP, Bilic G, Buhring HJ, Evangelista M, et al. . Concise review: isolation and characterization of cells from human term placenta: outcome of the first international Workshop on Placenta Derived Stem Cells. Stem Cells. (2008) 26:1–11. 10.1634/stemcells.2007-0594 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

