Proline Biosynthesis Enzyme Genes Confer Salt Tolerance to Switchgrass (Panicum virgatum L.) in Cooperation With Polyamines Metabolism
- PMID: 32117384
- PMCID: PMC7033549
- DOI: 10.3389/fpls.2020.00046
Proline Biosynthesis Enzyme Genes Confer Salt Tolerance to Switchgrass (Panicum virgatum L.) in Cooperation With Polyamines Metabolism
Abstract
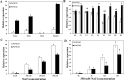
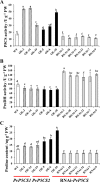
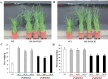
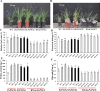
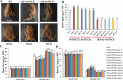
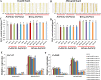
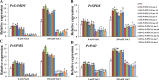
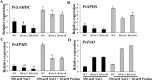
Understanding the regulation of proline metabolism necessitates the suppression of two Δ1-pyrroline-5-carboxylate synthetase enzyme (P5CS) genes performed in switchgrass (Panicum virgatum L.). The results reveal that overexpressing PvP5CS1 and PvP5CS2 increased salt tolerance. Additionally, transcript levels of spermidine (Spd) and spermine (Spm) synthesis and metabolism related genes were upregulated in PvP5CS OE-transgenic plants and downregulated in the PvP5CS RNAi transformants. According to salt stress assay and the measurement of transcript levels of Polyamines (PAs) metabolism-related genes, P5CS enzyme may not only be the key regulator of proline biosynthesis in switchgrass, but it may also indirectly affect the entire subset of pathway for ornithine to proline or to putrescine (Put). Furthermore, application of proline prompted expression levels of Spd and Spm synthesis and metabolism-related genes in both PvP5CS-RNAi and WT plants, but transcript levels were even lower in PvP5CS-RNAi compared to WT plants under salt stress condition. These results suggested that exogenous proline could accelerate polyamines metabolisms under salt stress. Nevertheless, the enzymes involved in this process and the potential functions remain poorly understood. Thus, the aim of this study is to reveal how proline functions with PAs metabolism under salt stress in switchgrass.
Keywords: PvP5CS1 and PvP5CS2; polyamines; proline; salt stress; switchgrass.
Copyright © 2020 Guan, Cui, Liu, Li, Li and Zhang.
Figures
Similar articles
-
ADP-ribosylation factors improve biomass yield and salinity tolerance in transgenic switchgrass (Panicum virgatum L.).Plant Cell Rep. 2020 Dec;39(12):1623-1638. doi: 10.1007/s00299-020-02589-x. Epub 2020 Sep 3. Plant Cell Rep. 2020. PMID: 32885306
-
Two P5CS genes from common bean exhibiting different tolerance to salt stress in transgenic Arabidopsis.J Genet. 2013 Dec;92(3):461-9. doi: 10.1007/s12041-013-0292-5. J Genet. 2013. PMID: 24371167
-
Overexpression of gene encoding the key enzyme involved in proline-biosynthesis (PuP5CS) to improve salt tolerance in switchgrass (Panicum virgatum L.).Plant Cell Rep. 2018 Aug;37(8):1187-1199. doi: 10.1007/s00299-018-2304-7. Epub 2018 May 25. Plant Cell Rep. 2018. PMID: 29802436
-
Regulation of levels of proline as an osmolyte in plants under water stress.Plant Cell Physiol. 1997 Oct;38(10):1095-102. doi: 10.1093/oxfordjournals.pcp.a029093. Plant Cell Physiol. 1997. PMID: 9399433 Review.
-
Molecular evolution of plant P5CS gene involved in proline biosynthesis.Mol Biol Rep. 2013 Nov;40(11):6429-35. doi: 10.1007/s11033-013-2757-2. Epub 2013 Sep 26. Mol Biol Rep. 2013. PMID: 24068435 Review.
Cited by
-
Dynamic transcriptomics and physiological insights reveal multi-tissue salt adaptation mechanisms in Amaranthus hypochondriacus across stress gradients.Plant Cell Rep. 2025 May 3;44(5):111. doi: 10.1007/s00299-025-03506-w. Plant Cell Rep. 2025. PMID: 40317365
-
Characterization of Differentially Expressed Genes under Salt Stress in Olive.Int J Mol Sci. 2021 Dec 23;23(1):154. doi: 10.3390/ijms23010154. Int J Mol Sci. 2021. PMID: 35008580 Free PMC article.
-
24-Epibrassinolide Reduces Drought-Induced Oxidative Stress by Modulating the Antioxidant System and Respiration in Wheat Seedlings.Plants (Basel). 2024 Jan 5;13(2):148. doi: 10.3390/plants13020148. Plants (Basel). 2024. PMID: 38256702 Free PMC article.
-
Tolerance Mechanisms of Olive Tree (Olea europaea) under Saline Conditions.Plants (Basel). 2024 Jul 29;13(15):2094. doi: 10.3390/plants13152094. Plants (Basel). 2024. PMID: 39124213 Free PMC article. Review.
-
Abiotic Stress in Crop Production.Int J Mol Sci. 2023 Apr 1;24(7):6603. doi: 10.3390/ijms24076603. Int J Mol Sci. 2023. PMID: 37047573 Free PMC article. Review.
References
-
- Bates L. S., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. 10.1007/BF00018060 - DOI
-
- Bouchereau A., Aziz A., Larher F., Martin-Tanguy J. (1999). Polyamines and environmental challenges: recent development. Plant Sci. 140 (2), 103–125. 10.1016/S0168-9452(98)00218-0 - DOI
-
- Casler M. D., Vogel K. P., Taliaferro C. M., Ehlke N. J., Berdahl J. D., Brummer E. C., et al. (2007). Latitudinal and longitudinal adaptation of switchgrass populations. Crop Sci. 47 (6), 2249–2260. 10.2135/cropsci2006.12.0780 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

