H3K9 Demethylation-Induced R-Loop Accumulation Is Linked to Disorganized Nucleoli
- PMID: 32117455
- PMCID: PMC7025566
- DOI: 10.3389/fgene.2020.00043
H3K9 Demethylation-Induced R-Loop Accumulation Is Linked to Disorganized Nucleoli
Abstract
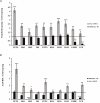
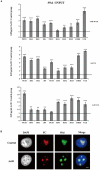
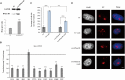
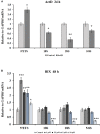
The nucleolar structure and integrity are important for a range of cellular functions of the nucleoli. It has been shown that cells lacking histone H3 Lysine 9 (H3K9) methylation form fragmented nucleoli. However, the molecular mechanism involved remains poorly understood. Here, we present evidence suggesting that loss of H3K9 dimethylation (H3K9me2) triggers R-loop accumulation at the rDNA locus, which further leads to the multilobed nucleoli. We reveal that suppression of H3K9 methyltransferase G9a by the inhibitor BIX 01294 causes R-loop accumulation at the rDNA region as well as inducing formation of multiple nucleoli. SiRNA-mediated knockdown of RNase H1 which can hydrolyze the RNA chain in R-loops causes an increase in R-loop formation, which in turn results in multiple nucleoli in one nucleus, whereas H3K9me2 levels are not affected by R-loop accumulation. Inhibition of RNA polymerase I transcription elongation by small molecule inhibitors induces a substantial decrease in H3K9me2 levels, accumulation of R-loops at rDNA sites, and nucleolus fragmentation. These results provide a mechanistic insight into the role of H3K9me2 in the structural integrity and organization of nucleoli via regulating R-loop accumulation.
Keywords: H3K9 dimethylation; R-loops; multiple nucleoli; ribosomal DNA; transcription inhibition.
Copyright © 2020 Zhou, Li, Wang, Hu, Zhao, Gautam and Li.
Figures





References
LinkOut - more resources
Full Text Sources

