Circular RNA hsa_circ_0084443 Is Upregulated in Diabetic Foot Ulcer and Modulates Keratinocyte Migration and Proliferation
- PMID: 32117579
- PMCID: PMC7047102
- DOI: 10.1089/wound.2019.0956
Circular RNA hsa_circ_0084443 Is Upregulated in Diabetic Foot Ulcer and Modulates Keratinocyte Migration and Proliferation
Abstract
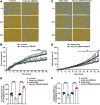
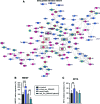
Objective: Insufficient knowledge about the molecular pathology of diabetic foot ulcer (DFU) impedes the development of effective wound treatment. Circular RNAs (circRNAs) are a novel class of RNA recently discovered to be widely expressed and have important biological functions; however, their role in skin wound healing remains largely unexplored. In this study, we investigated the role of circRNAs in DFU. Approach: CircRNA expression was profiled in normal wounds (NWs) and DFUs by microarray analysis, and hsa_circ_0084443 was identified as differentially expressed. The circularity and subcellular localization of hsa_circ_0084443 were characterized by northern blotting, real-time PCR, and fluorescence in situ hybridization. Cell migration, cell growth, and the transcriptome of human primary keratinocytes were analyzed after overexpression or RNA interference of hsa_circ_0084443. Results: hsa_circ_0084443 is downregulated in NWs compared with intact skin, and its level is higher in DFUs than NWs. We confirmed its circularity and presence in the cytoplasm of human epidermal keratinocytes. We showed that hsa_circ_0084443 reduced motility while enhancing the growth of keratinocytes. Furthermore, we identified a gene network with the potential to mediate the biological effect of hsa_circ_0084443. Innovation: CircRNAs have a functional role and a potential clinical significance in skin wound healing. Conclusions: We identified hsa_circ_0084443, a circRNA downregulated during NW healing, as a negative regulator of keratinocyte migration. Higher levels of hsa_circ_0084443 were detected in DFU samples, suggesting that it plays a role in pathology. These findings pave the way to understanding the functional role of circRNAs in human skin wound healing.
Keywords: circular RNA; diabetic foot ulcer; keratinocyte; noncoding RNA; wound healing.
Copyright © 2020 by Mary Ann Liebert, Inc., publishers.
Conflict of interest statement
No competing financial interests exist. The content of this article was expressly written by the authors listed. No ghostwriters were used to write this article.
Figures






References
-
- World Health Organization. Global Report on Diabetes. Geneva, Switzerland: World Health Organization, 2016;978:88
-
- Frykberg RG, Zgonis T, Armstrong DG, et al. Diabetic foot disorders. A clinical practice guideline (2006 revision). J Foot Ankle Surg 2006;45:S1–S66 - PubMed
-
- Baltzis D, Eleftheriadou I, Veves A. Pathogenesis and treatment of impaired wound healing in diabetes mellitus: new insights. Adv Ther 2014;31:817–836 - PubMed
-
- Catrina SB, Zheng X. Disturbed hypoxic responses as a pathogenic mechanism of diabetic foot ulcers. Diabetes Metab Res Rev 2016;32:179–185 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

