Supercoiling, R-loops, Replication and the Functions of Bacterial Type 1A Topoisomerases
- PMID: 32120891
- PMCID: PMC7140829
- DOI: 10.3390/genes11030249
Supercoiling, R-loops, Replication and the Functions of Bacterial Type 1A Topoisomerases
Abstract
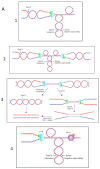
Type 1A topoisomerases (topos) are the only topos that bind single-stranded DNA and the only ones found in all cells of the three domains of life. Two subfamilies, topo I and topo III, are present in bacteria. Topo I, found in all of them, relaxes negative supercoiling, while topo III acts as a decatenase in replication. However, recent results suggest that they can also act as back-up for each other. Because they are ubiquitous, type 1A enzymes are expected to be essential for cell viability. Single topA (topo I) and topB (topo III) null mutants of Escherichia coli are viable, but for topA only with compensatory mutations. Double topA topB null mutants were initially believed to be non-viable. However, in two independent studies, results of next generation sequencing (NGS) have recently shown that double topA topB null mutants of Bacillus subtilis and E. coli are viable when they carry parC parE gene amplifications. These genes encode the two subunits of topo IV, the main cellular decatenase. Here, we discuss the essential functions of bacterial type 1A topos in the context of this observation and new results showing their involvement in preventing unregulated replication from R-loops.
Keywords: PriA; R-loop; oriC; replication; supercoiling; topA; topB; topoisomerase I; topoisomerase III; topoisomerases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Garnier F., Debat H., Nadal M. Type IA DNA Topoisomerases: A Universal Core and Multiple Activities. Methods Mol. Biol. 2018;1703:1–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

