Histone Post-Translational Modifications and CircRNAs in Mouse and Human Spermatozoa: Potential Epigenetic Marks to Assess Human Sperm Quality
- PMID: 32121034
- PMCID: PMC7141194
- DOI: 10.3390/jcm9030640
Histone Post-Translational Modifications and CircRNAs in Mouse and Human Spermatozoa: Potential Epigenetic Marks to Assess Human Sperm Quality
Abstract
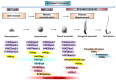
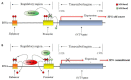
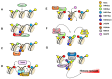
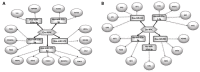
Spermatozoa (SPZ) are motile cells, characterized by a cargo of epigenetic information including histone post-translational modifications (histone PTMs) and non-coding RNAs. Specific histone PTMs are present in developing germ cells, with a key role in spermatogenic events such as self-renewal and commitment of spermatogonia (SPG), meiotic recombination, nuclear condensation in spermatids (SPT). Nuclear condensation is related to chromatin remodeling events and requires a massive histone-to-protamine exchange. After this event a small percentage of chromatin is condensed by histones and SPZ contain nucleoprotamines and a small fraction of nucleohistone chromatin carrying a landascape of histone PTMs. Circular RNAs (circRNAs), a new class of non-coding RNAs, characterized by a nonlinear back-spliced junction, able to play as microRNA (miRNA) sponges, protein scaffolds and translation templates, have been recently characterized in both human and mouse SPZ. Since their abundance in eukaryote tissues, it is challenging to deepen their biological function, especially in the field of reproduction. Here we review the critical role of histone PTMs in male germ cells and the profile of circRNAs in mouse and human SPZ. Furthermore, we discuss their suggested role as novel epigenetic biomarkers to assess sperm quality and improve artificial insemination procedure.
Keywords: circular RNAs (circRNAs); embryo development; fertilization; histone post-translational modifications (histone PTMs); human male infertility; sperm; spermatogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chianese R., Pierantoni R. Sperm Cells, an efficient shuttle for the intergenerational epigenetic memory. J. Pituit. Res. Treat. 2016;1:e001.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

