Molecular Evolution and Diversification of Proteins Involved in miRNA Maturation Pathway
- PMID: 32121542
- PMCID: PMC7154892
- DOI: 10.3390/plants9030299
Molecular Evolution and Diversification of Proteins Involved in miRNA Maturation Pathway
Abstract
Small RNAs (smRNA, 19-25 nucleotides long), which are transcribed by RNA polymerase II, regulate the expression of genes involved in a multitude of processes in eukaryotes. miRNA biogenesis and the proteins involved in the biogenesis pathway differ across plant and animal lineages. The major proteins constituting the biogenesis pathway, namely, the Dicers (DCL/DCR) and Argonautes (AGOs), have been extensively studied. However, the accessory proteins (DAWDLE (DDL), SERRATE (SE), and TOUGH (TGH)) of the pathway that differs across the two lineages remain largely uncharacterized. We present the first detailed report on the molecular evolution and divergence of these proteins across eukaryotes. Although DDL is present in eukaryotes and prokaryotes, SE and TGH appear to be specific to eukaryotes. The addition/deletion of specific domains and/or domain-specific sequence divergence in the three proteins points to the observed functional divergence of these proteins across the two lineages, which correlates with the differences in miRNA length across the two lineages. Our data enhance the current understanding of the structure-function relationship of these proteins and reveals previous unexplored crucial residues in the three proteins that can be used as a basis for further functional characterization. The data presented here on the number of miRNAs in crown eukaryotic lineages are consistent with the notion of the expansion of the number of miRNA-coding genes in animal and plant lineages correlating with organismal complexity. Whether this difference in functionally correlates with the diversification (or presence/absence) of the three proteins studied here or the miRNA signaling in the plant and animal lineages is unclear. Based on our results of the three proteins studied here and previously available data concerning the evolution of miRNA genes in the plant and animal lineages, we believe that miRNAs probably evolved once in the ancestor to crown eukaryotes and have diversified independently in the eukaryotes.
Keywords: Dawdle (DDL); Tough (TGH), Serrate (SE/ARS2), Argonaute (AGO), Dicer-Like (DCR/DCL), evolution; phylogeny; small RNA (smRNAs).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




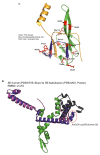
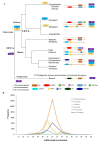
References
Grants and funding
LinkOut - more resources
Full Text Sources

