The First Plastid Genome of the Holoparasitic Genus Prosopanche (Hydnoraceae)
- PMID: 32121567
- PMCID: PMC7154897
- DOI: 10.3390/plants9030306
The First Plastid Genome of the Holoparasitic Genus Prosopanche (Hydnoraceae)
Abstract
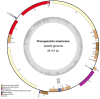
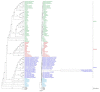
Plastomes of parasitic and mycoheterotrophic plants show different degrees of reduction depending on the plants' level of heterotrophy and host dependence in comparison to photoautotrophic sister species, and the amount of time since heterotrophic dependence was established. In all but the most recent heterotrophic lineages, this reduction involves substantial decrease in genome size and gene content and sometimes alterations of genome structure. Here, we present the first plastid genome of the holoparasitic genus Prosopanche, which shows clear signs of functionality. The plastome of Prosopanche americana has a length of 28,191 bp and contains only 24 unique genes, i.e., 14 ribosomal protein genes, four ribosomal RNA genes, five genes coding for tRNAs and three genes with other or unknown function (accD, ycf1, ycf2). The inverted repeat has been lost. Despite the split of Prosopanche and Hydnora about 54 MYA ago, the level of genome reduction is strikingly congruent between the two holoparasites although highly dissimilar nucleotide sequences are observed. Our results lead to two possible evolutionary scenarios that will be tested in the future with a larger sampling: 1) a Hydnoraceae plastome, similar to those of Hydnora and Prosopanche today, existed already in the most recent common ancestor and has not changed much with respect to gene content and structure, or 2) the genome similarities we observe today are the result of two independent evolutionary trajectories leading to almost the same endpoint. The first hypothesis would be most parsimonious whereas the second would point to taxon dependent essential gene sets for plants released from photosynthetic constraints.
Keywords: Hydnora; Hydnoraceae; Piperales; Prosopanche; holoparasite; parasitic plants; plastid genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Ohyama K., Fukuzawa H., Kohchi T., Shirai H., Sano T., Sano S., Umesono K., Shiki Y., Takeuchi Y., Chang Z., et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572. doi: 10.1038/322572a0. - DOI
-
- Wicke S., Müller K.F., de Pamphilis C.W., Quandt D., Wickett N.J., Zhang Y., Renner S.S., Schneeweiss G.M. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25:3711–3725. doi: 10.1105/tpc.113.113373. - DOI - PMC - PubMed
-
- Wicke S., Naumann J. Advances in Botanical Research. Volume 85. Academic Press; Cambridge, MA, USA: 2018. Molecular evolution of plastid genomes in parasitic flowering plants; pp. 315–347.
LinkOut - more resources
Full Text Sources

