Cell-penetrating peptide inhibits retromer-mediated human papillomavirus trafficking during virus entry
- PMID: 32123072
- PMCID: PMC7084110
- DOI: 10.1073/pnas.1917748117
Cell-penetrating peptide inhibits retromer-mediated human papillomavirus trafficking during virus entry
Abstract
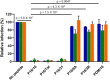
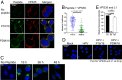
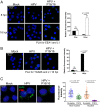
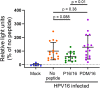
Virus replication requires critical interactions between viral proteins and cellular proteins that mediate many aspects of infection, including the transport of viral genomes to the site of replication. In human papillomavirus (HPV) infection, the cellular protein complex known as retromer binds to the L2 capsid protein and sorts incoming virions into the retrograde transport pathway for trafficking to the nucleus. Here, we show that short synthetic peptides containing the HPV16 L2 retromer-binding site and a cell-penetrating sequence enter cells, sequester retromer from the incoming HPV pseudovirus, and inhibit HPV exit from the endosome, resulting in loss of viral components from cells and in a profound, dose-dependent block to infection. The peptide also inhibits cervicovaginal HPV16 pseudovirus infection in a mouse model. These results confirm the retromer-mediated model of retrograde HPV entry and validate intracellular virus trafficking as an antiviral target. More generally, inhibiting virus replication with agents that can enter cells and disrupt essential protein-protein interactions may be applicable in broad outline to many viruses.
Keywords: HPV; antiviral agent; protein transduction domain; retrograde; retromer.
Conflict of interest statement
Competing interest statement: P.Z. and D.D. are inventors on a patent application related to this work.
Figures


Similar articles
-
Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection.PLoS Pathog. 2015 Feb 18;11(2):e1004699. doi: 10.1371/journal.ppat.1004699. eCollection 2015 Feb. PLoS Pathog. 2015. PMID: 25693203 Free PMC article.
-
Cell-Penetrating Peptide Mediates Intracellular Membrane Passage of Human Papillomavirus L2 Protein to Trigger Retrograde Trafficking.Cell. 2018 Sep 6;174(6):1465-1476.e13. doi: 10.1016/j.cell.2018.07.031. Epub 2018 Aug 16. Cell. 2018. PMID: 30122350 Free PMC article.
-
Efficient Inhibition of Human Papillomavirus Infection by L2 Minor Capsid-Derived Lipopeptide.mBio. 2019 Aug 6;10(4):e01834-19. doi: 10.1128/mBio.01834-19. mBio. 2019. PMID: 31387913 Free PMC article.
-
Subcellular Trafficking of the Papillomavirus Genome during Initial Infection: The Remarkable Abilities of Minor Capsid Protein L2.Viruses. 2017 Dec 3;9(12):370. doi: 10.3390/v9120370. Viruses. 2017. PMID: 29207511 Free PMC article. Review.
-
L2, the minor capsid protein of papillomavirus.Virology. 2013 Oct;445(1-2):175-86. doi: 10.1016/j.virol.2013.04.017. Epub 2013 May 17. Virology. 2013. PMID: 23689062 Free PMC article. Review.
Cited by
-
Cell-Penetrating Antimicrobial Peptides with Anti-Infective Activity against Intracellular Pathogens.Antibiotics (Basel). 2022 Dec 8;11(12):1772. doi: 10.3390/antibiotics11121772. Antibiotics (Basel). 2022. PMID: 36551429 Free PMC article. Review.
-
Biological Membrane-Penetrating Peptides: Computational Prediction and Applications.Front Cell Infect Microbiol. 2022 Mar 25;12:838259. doi: 10.3389/fcimb.2022.838259. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402305 Free PMC article. Review.
-
Application of Cell Penetrating Peptides as a Promising Drug Carrier to Combat Viral Infections.Mol Biotechnol. 2023 Sep;65(9):1387-1402. doi: 10.1007/s12033-023-00679-1. Epub 2023 Jan 31. Mol Biotechnol. 2023. PMID: 36719639 Free PMC article. Review.
-
Analysis of HPV prevalence among individuals with reproductive tract infections in a Chinese population.Medicine (Baltimore). 2023 Oct 13;102(41):e34989. doi: 10.1097/MD.0000000000034989. Medicine (Baltimore). 2023. PMID: 37832113 Free PMC article.
-
Biotherapeutic effect of cell-penetrating peptides against microbial agents: a review.Tissue Barriers. 2022 Jul 3;10(3):1995285. doi: 10.1080/21688370.2021.1995285. Epub 2021 Oct 25. Tissue Barriers. 2022. PMID: 34694961 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

