Conformational control of small GTPases by AMPylation
- PMID: 32123090
- PMCID: PMC7084064
- DOI: 10.1073/pnas.1917549117
Conformational control of small GTPases by AMPylation
Abstract
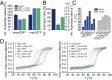
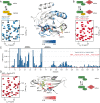
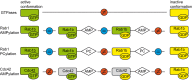
Posttranslational modifications (PTMs) are important physiological means to regulate the activities and structures of central regulatory proteins in health and disease. Small GTPases have been recognized as important molecules that are targeted by PTMs during infections of mammalian cells by bacterial pathogens. The enzymes DrrA/SidM and AnkX from Legionella pneumophila AMPylate and phosphocholinate Rab1b during infection, respectively. Cdc42 is AMPylated by IbpA from Histophilus somni at tyrosine 32 or by VopS from Vibrio parahaemolyticus at threonine 35. These modifications take place in the important regulatory switch I or switch II regions of the GTPases. Since Rab1b and Cdc42 are central regulators of intracellular vesicular trafficking and of the actin cytoskeleton, their modifications by bacterial pathogens have a profound impact on the course of infection. Here, we addressed the biochemical and structural consequences of GTPase AMPylation and phosphocholination. By combining biochemical experiments and NMR analysis, we demonstrate that AMPylation can overrule the activity state of Rab1b that is commonly dictated by binding to guanosine diphosphate or guanosine triphosphate. Thus, PTMs may exert conformational control over small GTPases and may add another previously unrecognized layer of activity control to this important regulatory protein family.
Keywords: NMR; posttranslational modifications; protein dynamics; small GTPases.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Traut T. W., Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 (1994). - PubMed
-
- Cherfils J., Zeghouf M., Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 (2013). - PubMed
-
- Zoppino F. C. M., Militello R. D., Slavin I., Alvarez C., Colombo M. I., Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11, 1246–1261 (2010). - PubMed
-
- Dong N., et al. , Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150, 1029–1041 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

