bioPROTACs as versatile modulators of intracellular therapeutic targets including proliferating cell nuclear antigen (PCNA)
- PMID: 32123106
- PMCID: PMC7084165
- DOI: 10.1073/pnas.1920251117
bioPROTACs as versatile modulators of intracellular therapeutic targets including proliferating cell nuclear antigen (PCNA)
Abstract
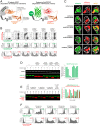
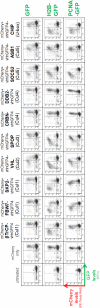
Targeted degradation approaches such as proteolysis targeting chimeras (PROTACs) offer new ways to address disease through tackling challenging targets and with greater potency, efficacy, and specificity over traditional approaches. However, identification of high-affinity ligands to serve as PROTAC starting points remains challenging. As a complementary approach, we describe a class of molecules termed biological PROTACs (bioPROTACs)-engineered intracellular proteins consisting of a target-binding domain directly fused to an E3 ubiquitin ligase. Using GFP-tagged proteins as model substrates, we show that there is considerable flexibility in both the choice of substrate binders (binding positions, scaffold-class) and the E3 ligases. We then identified a highly effective bioPROTAC against an oncology target, proliferating cell nuclear antigen (PCNA) to elicit rapid and robust PCNA degradation and associated effects on DNA synthesis and cell cycle progression. Overall, bioPROTACs are powerful tools for interrogating degradation approaches, target biology, and potentially for making therapeutic impacts.
Keywords: PCNA; bioPROTAC; targeted degradation.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

