Lactate dehydrogenase inhibition synergizes with IL-21 to promote CD8+ T cell stemness and antitumor immunity
- PMID: 32123114
- PMCID: PMC7084161
- DOI: 10.1073/pnas.1920413117
Lactate dehydrogenase inhibition synergizes with IL-21 to promote CD8+ T cell stemness and antitumor immunity
Abstract
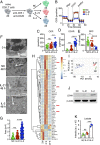
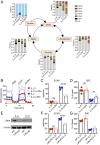
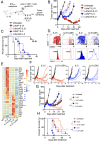
Interleukin (IL)-2 and IL-21 dichotomously shape CD8+ T cell differentiation. IL-2 drives terminal differentiation, generating cells that are poorly effective against tumors, whereas IL-21 promotes stem cell memory T cells (TSCM) and antitumor responses. Here we investigated the role of metabolic programming in the developmental differences induced by these cytokines. IL-2 promoted effector-like metabolism and aerobic glycolysis, robustly inducing lactate dehydrogenase (LDH) and lactate production, whereas IL-21 maintained a metabolically quiescent state dependent on oxidative phosphorylation. LDH inhibition rewired IL-2-induced effects, promoting pyruvate entry into the tricarboxylic acid cycle and inhibiting terminal effector and exhaustion programs, including mRNA expression of members of the NR4A family of nuclear receptors, as well as Prdm1 and Xbp1 While deletion of Ldha prevented development of cells with antitumor effector function, transient LDH inhibition enhanced the generation of memory cells capable of triggering robust antitumor responses after adoptive transfer. LDH inhibition did not significantly affect IL-21-induced metabolism but caused major transcriptomic changes, including the suppression of IL-21-induced exhaustion markers LAG3, PD1, 2B4, and TIM3. LDH inhibition combined with IL-21 increased the formation of TSCM cells, resulting in more profound antitumor responses and prolonged host survival. These findings indicate a pivotal role for LDH in modulating cytokine-mediated T cell differentiation and underscore the therapeutic potential of transiently inhibiting LDH during adoptive T cell-based immunotherapy, with an unanticipated cooperative antitumor effect of LDH inhibition and IL-21.
Keywords: IL-2; IL-21; LDH; adoptive immunotherapy; immunometabolism.
Conflict of interest statement
Competing interest statement: W.J.L. is an inventor on patents or patent applications related to IL-2 and IL-21. D.H., S.G., L.M.N., L.G., and W.J.L. are inventors on patent application(s) related to the LDH inhibitor described herein. L.G. is an inventor on a patent related to methods for generating TSCM cells.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

