The crosstalk of NAD, ROS and autophagy in cellular health and ageing
- PMID: 32124104
- PMCID: PMC7196094
- DOI: 10.1007/s10522-020-09864-0
The crosstalk of NAD, ROS and autophagy in cellular health and ageing
Abstract
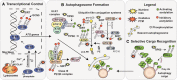
Cellular adaptation to various types of stress requires a complex network of steps that altogether lead to reconstitution of redox balance, degradation of damaged macromolecules and restoration of cellular metabolism. Advances in our understanding of the interplay between cellular signalling and signal translation paint a complex picture of multi-layered paths of regulation. In this review we explore the link between cellular adaptation to metabolic and oxidative stresses by activation of autophagy, a crucial cellular catabolic pathway. Metabolic stress can lead to changes in the redox state of nicotinamide adenine dinucleotide (NAD), a co-factor in a variety of enzymatic reactions and thus trigger autophagy that acts to sequester intracellular components for recycling to support cellular growth. Likewise, autophagy is activated by oxidative stress to selectively recycle damaged macromolecules and organelles and thus maintain cellular viability. Multiple proteins that help regulate or execute autophagy are targets of post-translational modifications (PTMs) that have an effect on their localization, binding affinity or enzymatic activity. These PTMs include acetylation, a reversible enzymatic modification of a protein's lysine residues, and oxidation, a set of reversible and irreversible modifications by free radicals. Here we highlight the latest findings and outstanding questions on the interplay of autophagy with metabolic stress, presenting as changes in NAD levels, and oxidative stress, with a focus on autophagy proteins that are regulated by both, oxidation and acetylation. We further explore the relevance of this multi-layered signalling to healthy human ageing and their potential role in human disease.
Keywords: Acetylation; Ageing; Autophagy; NAD; ROS; Sirtuins.
Figures

References
-
- Ahmad S, et al. Protein oxidation: an overview of metabolism of sulphur containing amino acid, cysteine. Front Biosci (Schol Ed) 2017;9:71–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

