PIK3Cδ expression by fibroblasts promotes triple-negative breast cancer progression
- PMID: 32125284
- PMCID: PMC7260014
- DOI: 10.1172/JCI128313
PIK3Cδ expression by fibroblasts promotes triple-negative breast cancer progression
Abstract
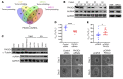
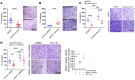
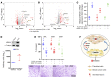
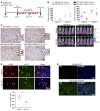
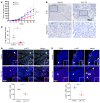
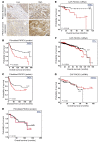
As there is growing evidence for the tumor microenvironment's role in tumorigenesis, we investigated the role of fibroblast-expressed kinases in triple-negative breast cancer (TNBC). Using a high-throughput kinome screen combined with 3D invasion assays, we identified fibroblast-expressed PIK3Cδ (f-PIK3Cδ) as a key regulator of cancer progression. Although PIK3Cδ was expressed in primary fibroblasts derived from TNBC patients, it was barely detectable in breast cancer (BC) cell lines. Genetic and pharmacological gain- and loss-of-function experiments verified the contribution of f-PIK3Cδ in TNBC cell invasion. Integrated secretomics and transcriptomics analyses revealed a paracrine mechanism via which f-PIK3Cδ confers its protumorigenic effects. Inhibition of f-PIK3Cδ promoted the secretion of factors, including PLGF and BDNF, that led to upregulation of NR4A1 in TNBC cells, where it acts as a tumor suppressor. Inhibition of PIK3Cδ in an orthotopic BC mouse model reduced tumor growth only after inoculation with fibroblasts, indicating a role of f-PIK3Cδ in cancer progression. Similar results were observed in the MMTV-PyMT transgenic BC mouse model, along with a decrease in tumor metastasis, emphasizing the potential immune-independent effects of PIK3Cδ inhibition. Finally, analysis of BC patient cohorts and TCGA data sets identified f-PIK3Cδ (protein and mRNA levels) as an independent prognostic factor for overall and disease-free survival, highlighting it as a therapeutic target for TNBC.
Keywords: Breast cancer; Cell Biology; Oncology; Protein kinases; Signal transduction.
Conflict of interest statement
Figures



References
-
- Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109(9):1721–1728. doi: 10.1002/cncr.22618. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

