MicroRNA-146a regulates immune-related adverse events caused by immune checkpoint inhibitors
- PMID: 32125286
- PMCID: PMC7213806
- DOI: 10.1172/jci.insight.132334
MicroRNA-146a regulates immune-related adverse events caused by immune checkpoint inhibitors
Abstract
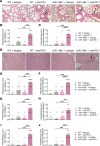
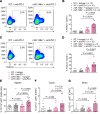
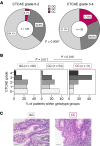
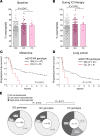
Immune checkpoint inhibitor (ICI) therapy has shown a significant benefit in the treatment of a variety of cancer entities. However, immune-related adverse events (irAEs) occur frequently and can lead to ICI treatment termination. MicroRNA-146a (miR-146a) has regulatory functions in immune cells. We observed that mice lacking miR-146a developed markedly more severe irAEs compared with WT mice in several irAE target organs in 2 different murine models. miR-146a-/- mice exhibited increased T cell activation and effector function upon ICI treatment. Moreover, neutrophil numbers in the spleen and the inflamed intestine were highly increased in ICI-treated miR-146a-/- mice. Therapeutic administration of a miR-146a mimic reduced irAE severity. To validate our preclinical findings in patients, we analyzed the effect of a SNP in the MIR146A gene on irAE severity in 167 patients treated with ICIs. We found that the SNP rs2910164 leading to reduced miR-146a expression was associated with an increased risk of developing severe irAEs, reduced progression-free survival, and increased neutrophil counts both at baseline and during ICI therapy. In conclusion, we characterized miR-146a as a molecular target for preventing ICI-mediated autoimmune dysregulation. Furthermore, we identified the MIR146A SNP rs2910164 as a biomarker to predict severe irAE development in ICI-treated patients.
Keywords: Adaptive immunity; Cancer immunotherapy; Inflammation; Noncoding RNAs; Oncology.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

