Quantitative proteomics to study aging in rabbit liver
- PMID: 32126221
- PMCID: PMC7138690
- DOI: 10.1016/j.mad.2020.111227
Quantitative proteomics to study aging in rabbit liver
Abstract
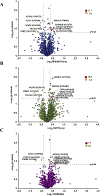
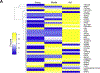
Aging globally effects cellular and organismal metabolism across a range of mammalian species, including humans and rabbits. Rabbits (Oryctolagus cuniculus are an attractive model system of aging due to their genetic similarity with humans and their short lifespans. This model can be used to understand metabolic changes in aging especially in major organs such as liver where we detected pronounced variations in fat metabolism, mitochondrial dysfunction, and protein degradation. Such changes in the liver are consistent across several mammalian species however in rabbits the downstream effects of these changes have not yet been explored. We have applied proteomics to study changes in the liver proteins from young, middle, and old age rabbits using a multiplexing cPILOT strategy. This resulted in the identification of 2,586 liver proteins, among which 45 proteins had significant p < 0.05) changes with aging. Seven proteins were differentially-expressed at all ages and include fatty acid binding protein, aldehyde dehydrogenase, enoyl-CoA hydratase, 3-hydroxyacyl CoA dehydrogenase, apolipoprotein C3, peroxisomal sarcosine oxidase, adhesion G-protein coupled receptor, and glutamate ionotropic receptor kinate. Insights to how alterations in metabolism affect protein expression in liver have been gained and demonstrate the utility of rabbit as a model of aging.
Keywords: Aging; Enhanced multiplexing; Liver; Metabolism; Oryctolagus cuniculus; Proteomics; Rabbit; cPILOT.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest None.
Figures
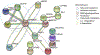

References
-
- Durani LW; Hamezah HS; Ibrahim NF; Yanagisawa D; Makpol S; Damanhuri HA; Tooyama I, Age-related changes in the metabolic profiles of rat hippocampus, medial prefrontal cortex and striatum. Biochem Biophys Res Commun 2017, 493 (3), 1356–1363. - PubMed
-
- Lee JS; Ward WO; Ren H; Vallanat B; Darlington GJ; Han ES; Laguna JC; DeFord JH; Papaconstantinou J; Selman C; Corton JC, Meta-analysis of gene expression in the mouse liver reveals biomarkers associated with inflammation increased early during aging. Mech Ageing Dev 2012, 133 (7), 467–78. - PubMed
-
- United Nations, D. o. E. a. S. A., Population Division World Population Ageing 2017 - Highlights United Nations: New York, USA, 2017; p (ST/ESA/SER.A/397).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

