Accurate targeted long-read DNA methylation and hydroxymethylation sequencing with TAPS
- PMID: 32127008
- PMCID: PMC7053107
- DOI: 10.1186/s13059-020-01969-6
Accurate targeted long-read DNA methylation and hydroxymethylation sequencing with TAPS
Abstract
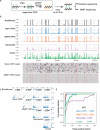
We present long-read Tet-assisted pyridine borane sequencing (lrTAPS) for targeted base-resolution sequencing of DNA methylation and hydroxymethylation in regions up to 10 kb from nanogram-level input. Compatible with both Oxford Nanopore and PacBio Single-Molecule Real-Time (SMRT) sequencing, lrTAPS detects methylation with accuracy comparable to short-read Illumina sequencing but with long-range epigenetic phasing. We applied lrTAPS to sequence difficult-to-map regions in mouse embryonic stem cells and to identify distinct methylation events in the integrated hepatitis B virus genome.
Keywords: 5-Methylcytosine; Bisulfite-free; DNA methylation; Epigenetic phasing; Long-read sequencing.
Conflict of interest statement
C-XS and YL are named as inventors on pending patent applications filed by the Ludwig Institute for Cancer Research pertaining to one or more aspects of the technologies described here, which have been licensed to Base Genomics.
Figures

References
-
- Wenger AM, Peluso P, Rowell WJ, Chang PC, Hall RJ, Concepcion GT, Ebler J, Fungtammasan A, Kolesnikov A, Olson ND, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37(10):1155–1162. doi: 10.1038/s41587-019-0217-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

