Targeting the E3 Ubiquitin Ligase PJA1 Enhances Tumor-Suppressing TGFβ Signaling
- PMID: 32127355
- PMCID: PMC7704101
- DOI: 10.1158/0008-5472.CAN-19-3116
Targeting the E3 Ubiquitin Ligase PJA1 Enhances Tumor-Suppressing TGFβ Signaling
Abstract
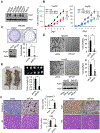
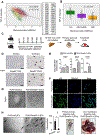
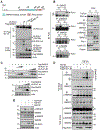
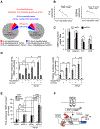
RING-finger E3 ligases are instrumental in the regulation of inflammatory cascades, apoptosis, and cancer. However, their roles are relatively unknown in TGFβ/SMAD signaling. SMAD3 and its adaptors, such as β2SP, are important mediators of TGFβ signaling and regulate gene expression to suppress stem cell-like phenotypes in diverse cancers, including hepatocellular carcinoma (HCC). Here, PJA1, an E3 ligase, promoted ubiquitination and degradation of phosphorylated SMAD3 and impaired a SMAD3/β2SP-dependent tumor-suppressing pathway in multiple HCC cell lines. In mice deficient for SMAD3 (Smad3 +/-), PJA1 overexpression promoted the transformation of liver stem cells. Analysis of genes regulated by PJA1 knockdown and TGFβ1 signaling revealed 1,584 co-upregulated genes and 1,280 co-downregulated genes, including many implicated in cancer. The E3 ligase inhibitor RTA405 enhanced SMAD3-regulated gene expression and reduced growth of HCC cells in culture and xenografts of HCC tumors, suggesting that inhibition of PJA1 may be beneficial in treating HCC or preventing HCC development in at-risk patients.Significance: These findings provide a novel mechanism regulating the tumor suppressor function of TGFβ in liver carcinogenesis.
©2020 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures
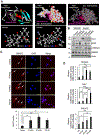
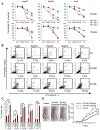
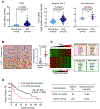
References
-
- International Agency for Research on Cancer: World Health Organization. GLOBOCAN 2014: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2014. . Lyon, France: International Agency for Research on Cancer: World Health Organization; [updated 2014; cited 2014 March 27] 2014:Available from: http://globocan.iarc.fr/Pages/fact_sheets_population.aspx.
-
- Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723–50 - PubMed
-
- Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15:599–616 - PubMed
-
- Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

