Bacterial Transformation Buffers Environmental Fluctuations through the Reversible Integration of Mobile Genetic Elements
- PMID: 32127449
- PMCID: PMC7064763
- DOI: 10.1128/mBio.02443-19
Bacterial Transformation Buffers Environmental Fluctuations through the Reversible Integration of Mobile Genetic Elements
Abstract
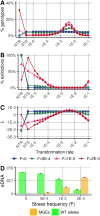
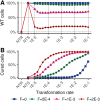
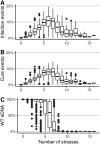
Horizontal gene transfer (HGT) promotes the spread of genes within bacterial communities. Among the HGT mechanisms, natural transformation stands out as being encoded by the bacterial core genome. Natural transformation is often viewed as a way to acquire new genes and to generate genetic mixing within bacterial populations. Another recently proposed function is the curing of bacterial genomes of their infectious parasitic mobile genetic elements (MGEs). Here, we propose that these seemingly opposing theoretical points of view can be unified. Although costly for bacterial cells, MGEs can carry functions that are at points in time beneficial to bacteria under stressful conditions (e.g., antibiotic resistance genes). Using computational modeling, we show that, in stochastic environments, an intermediate transformation rate maximizes bacterial fitness by allowing the reversible integration of MGEs carrying resistance genes, although these MGEs are costly for host cell replication. Based on this dual function (MGE acquisition and removal), transformation would be a key mechanism for stabilizing the bacterial genome in the long term, and this would explain its striking conservation.IMPORTANCE Natural transformation is the acquisition, controlled by bacteria, of extracellular DNA and is one of the most common mechanisms of horizontal gene transfer, promoting the spread of resistance genes. However, its evolutionary function remains elusive, and two main roles have been proposed: (i) the new gene acquisition and genetic mixing within bacterial populations and (ii) the removal of infectious parasitic mobile genetic elements (MGEs). While the first one promotes genetic diversification, the other one promotes the removal of foreign DNA and thus genome stability, making these two functions apparently antagonistic. Using a computational model, we show that intermediate transformation rates, commonly observed in bacteria, allow the acquisition then removal of MGEs. The transient acquisition of costly MGEs with resistance genes maximizes bacterial fitness in environments with stochastic stress exposure. Thus, transformation would ensure both a strong dynamic of the bacterial genome in the short term and its long-term stabilization.
Keywords: horizontal gene transfer; mobile genetic elements; natural transformation; resistance genes; stochastic environment.
Copyright © 2020 Carvalho et al.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
