Using Colonization Assays and Comparative Genomics To Discover Symbiosis Behaviors and Factors in Vibrio fischeri
- PMID: 32127462
- PMCID: PMC7064787
- DOI: 10.1128/mBio.03407-19
Using Colonization Assays and Comparative Genomics To Discover Symbiosis Behaviors and Factors in Vibrio fischeri
Abstract
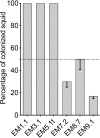
The luminous marine Gram-negative bacterium Vibrio (Aliivibrio) fischeri is the natural light organ symbiont of several squid species, including the Hawaiian bobtail squid, Euprymna scolopes, and the Japanese bobtail squid, Euprymna morsei Work with E. scolopes has shown how the bacteria establish their niche in the light organ of the newly hatched host. Two types of V. fischeri strains have been distinguished based upon their behavior in cocolonization competition assays in juvenile E. scolopes, i.e., (i) niche-sharing or (ii) niche-dominant behavior. This study aimed to determine whether these behaviors are observed with other V. fischeri strains or whether they are specific to those isolated from E. scolopes light organs. Cocolonization competition assays between V. fischeri strains isolated from the congeneric squid E. morsei or from other marine animals revealed the same sharing or dominant behaviors. In addition, whole-genome sequencing of these strains showed that the dominant behavior is polyphyletic and not associated with the presence or absence of a single gene or genes. Comparative genomics of 44 squid light organ isolates from around the globe led to the identification of symbiosis-specific candidates in the genomes of these strains. Colonization assays using genetic derivatives with deletions of these candidates established the importance of two such genes in colonization. This study has allowed us to expand the concept of distinct colonization behaviors to strains isolated from a number of squid and fish hosts.IMPORTANCE There is an increasing recognition of the importance of strain differences in the ecology of a symbiotic bacterial species and, in particular, how these differences underlie crucial interactions with their host. Nevertheless, little is known about the genetic bases for these differences, how they manifest themselves in specific behaviors, and their distribution among symbionts of different host species. In this study, we sequenced the genomes of Vibrio fischeri isolated from the tissues of squids and fishes and applied comparative genomics approaches to look for patterns between symbiont lineages and host colonization behavior. In addition, we identified the only two genes that were exclusively present in all V. fischeri strains isolated from the light organs of sepiolid squid species. Mutational studies of these genes indicated that they both played a role in colonization of the squid light organ, emphasizing the value of applying a comparative genomics approach in the study of symbioses.
Keywords: Aliivibrio; Vibrio (Aliivibrio) fischeri; Vibrio fischeri; dominance; genome analysis; genomes; intraspecific; symbiosis.
Copyright © 2020 Bongrand et al.
Figures






References
-
- Dunlap PV, Ast JC, Kimura S, Fukui A, Yoshino T, Endo H. 2007. Phylogenetic analysis of host-symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23:507–532. doi:10.1111/j.1096-0031.2007.00157.x. - DOI
-
- Kerwin AH, Gromek SM, Suria AM, Samples RM, Deoss DJ, O’Donnell K, Frasca S, Sutton DA, Wiederhold NP, Balunas MJ, Nyholm SV. 2019. Shielding the next generation: symbiotic bacteria from a reproductive organ protect bobtail squid eggs from fungal fouling. mBio 10:e02376–19. doi:10.1128/mBio.02376-19. - DOI - PMC - PubMed
-
- Nawroth JC, Guo H, Koch E, Heath-Heckman EAC, Hermanson JC, Ruby EG, Dabiri JO, Kanso E, McFall-Ngai M. 2017. Motile cilia create fluid-mechanical microhabitats for the active recruitment of the host microbiome. Proc Natl Acad Sci U S A 114:9510–9516. doi:10.1073/pnas.1706926114. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
