Frameshifting preserves key physicochemical properties of proteins
- PMID: 32127487
- PMCID: PMC7084103
- DOI: 10.1073/pnas.1911203117
Frameshifting preserves key physicochemical properties of proteins
Abstract
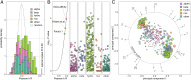
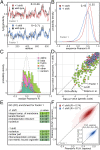
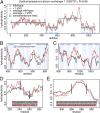
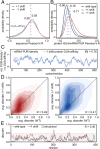
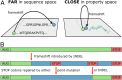
Frameshifts in protein coding sequences are widely perceived as resulting in either nonfunctional or even deleterious protein products. Indeed, frameshifts typically lead to markedly altered protein sequences and premature stop codons. By analyzing complete proteomes from all three domains of life, we demonstrate that, in contrast, several key physicochemical properties of protein sequences exhibit significant robustness against +1 and -1 frameshifts. In particular, we show that hydrophobicity profiles of many protein sequences remain largely invariant upon frameshifting. For example, over 2,900 human proteins exhibit a Pearson's correlation coefficient R between the hydrophobicity profiles of the original and the +1-frameshifted variants greater than 0.7, despite an average sequence identity between the two of only 6.5% in this group. We observe a similar effect for protein sequence profiles of affinity for certain nucleobases as well as protein sequence profiles of intrinsic disorder. Finally, analysis of significance and optimality demonstrates that frameshift stability is embedded in the structure of the universal genetic code and may have contributed to shaping it. Our results suggest that frameshifting may be a powerful evolutionary mechanism for creating new proteins with vastly different sequences, yet similar physicochemical properties to the proteins from which they originate.
Keywords: evolution; frameshift; genetic code; hydrophobicity.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Stenson P. D., et al. , Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 21, 577–581 (2003). - PubMed
-
- Garcia-Diaz M., Kunkel T. A., Mechanism of a genetic glissando: Structural biology of indel mutations. Trends Biochem. Sci. 31, 206–214 (2006). - PubMed
-
- Maki H., Origins of spontaneous mutations: Specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu. Rev. Genet. 36, 279–303 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

