Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata
- PMID: 32127603
- PMCID: PMC7054531
- DOI: 10.1038/s41598-020-60803-y
Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata
Abstract
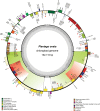
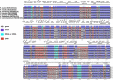
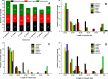
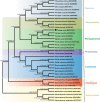
Plantago ovata (Plantaginaceae) is an economically and medicinally important species, however, least is known about its genomics and evolution. Here, we report the first complete plastome genome of P. ovata and comparison with previously published genomes of related species from Plantaginaceae. The results revealed that P. ovata plastome size was 162,116 bp and that it had typical quadripartite structure containing a large single copy region of 82,084 bp and small single copy region of 5,272 bp. The genome has a markedly higher inverted repeat (IR) size of 37.4 kb, suggesting large-scale inversion of 13.8 kb within the expanded IR regions. In addition, the P. ovata plastome contains 149 different genes, including 43 tRNA, 8 rRNA, and 98 protein-coding genes. The analysis revealed 139 microsatellites, of which 71 were in the non-coding regions. Approximately 32 forward, 34 tandem, and 17 palindromic repeats were detected. The complete genome sequences, 72 shared genes, matK gene, and rbcL gene from related species generated the same phylogenetic signals, and phylogenetic analysis revealed that P. ovata formed a single clade with P. maritima and P. media. The divergence time estimation as employed in BEAST revealed that P. ovata diverged from P. maritima and P. media about 11.0 million years ago (Mya; 95% highest posterior density, 10.06-12.25 Mya). In conclusion, P. ovata had significant variation in the IR region, suggesting a more stable P. ovata plastome genome than that of other Plantaginaceae species.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Palmer JD. Plastid chromosomes: structure and evolution. The molecular biology of plastids. 1991;7:5–53. doi: 10.1016/B978-0-12-715007-9.50009-8. - DOI
-
- Henry, R. J. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. (Cabi Publishing, 2005).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

