A Combined four-mRNA Signature Associated with Lymphatic Metastasis for Prognosis of Colorectal Cancer
- PMID: 32127941
- PMCID: PMC7052913
- DOI: 10.7150/jca.38796
A Combined four-mRNA Signature Associated with Lymphatic Metastasis for Prognosis of Colorectal Cancer
Abstract
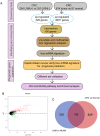
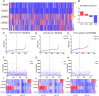
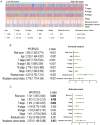
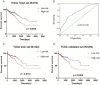
Background: Colorectal cancer (CRC) is one of the most common malignant tumors in the world. Lymph node metastasis (LNM) is a common mode of metastasis of CRC. However, the combined mRNA biomarkers associated with LNM of CRC that can effectively predict CRC prognosis have not been reported yet. Methods: To identify biomarkers that are associated with LNM, we collected data from the The Cancer Genome Atlas (TCGA) database. The edgeR package was searched to seek LNM-related genes by comparisons between cancer samples and normal colorectal tissues and between LNM and non-LNM (NLNM) of CRC. Univariate and multivariate regression analysis of genes in the intersection to build gene signature associated with independent prognosis of CRC, and then verified by Kaplan-Meier curve and log-rank test, receiver operating characteristic (ROC) curve was used to determine the efficiency of survival prediction of our four-mRNA signature. Finally, the potential molecular mechanisms and properties of these gene signature were also explored with functional and pathway enrichment analysis. Results: 329 mRNAs were up-regulated in CRC tissues with LNM, and 8461 mRNAs were up-regulated in CRC tissues, the intersection is 100 mRNAs. After univariate and multivariate Cox regression analysis of 100 mRNAs, a novel four LNM related mRNAs (EPHA8, KRT85, GABRA3, and CLPSL1) were screened as independent prognostic indicators of CRC. Surprisingly, the four-mRNA signature can predict the prognosis of CRC patients independently of clinical factors andthe area under the curve (AUC) of the ROC is 0.730. The novel four-mRNA signature was used to identify high and low-risk groups. Stratified analysis indicated the risk score based on four-mRNA signature was an independent prognostic indicator for female, T3+T4, N1+N2 ,stage III+IV and patients with no new tumor event. Functional annotation of this risk model in high-risk patients revealed that pathways associated with neuroactive ligand-receptor interaction, estrogen signaling pathway, and steroid hormone biosynthesis. Conclusions: By conducting TCGA data mining, our study demonstrated that a four-mRNA signature associated with LNM can be used as a combined biomarker for independent prognosis of CRC.
Keywords: biomarker; colorectal cancer; lymph node metastasis; mRNA; prognosis.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

Similar articles
-
Identification of critical prognosis signature associated with lymph node metastasis of stomach adenocarcinomas.World J Surg Oncol. 2023 Feb 23;21(1):61. doi: 10.1186/s12957-023-02940-y. World J Surg Oncol. 2023. PMID: 36823639 Free PMC article.
-
Esophageal cancer lymph node metastasis-associated gene signature optimizes overall survival prediction of esophageal cancer.J Cell Biochem. 2019 Jan;120(1):592-600. doi: 10.1002/jcb.27416. Epub 2018 Sep 22. J Cell Biochem. 2019. PMID: 30242875
-
Follistatin-Like 3 Correlates With Lymph Node Metastasis and Serves as a Biomarker of Extracellular Matrix Remodeling in Colorectal Cancer.Front Immunol. 2021 Jul 16;12:717505. doi: 10.3389/fimmu.2021.717505. eCollection 2021. Front Immunol. 2021. PMID: 34335633 Free PMC article.
-
A Five-microRNA Signature as Prognostic Biomarker in Colorectal Cancer by Bioinformatics Analysis.Front Oncol. 2019 Nov 12;9:1207. doi: 10.3389/fonc.2019.01207. eCollection 2019. Front Oncol. 2019. PMID: 31799184 Free PMC article.
-
The Application of Gene Expression Profiling in Predictions of Occult Lymph Node Metastasis in Colorectal Cancer Patients.Biomedicines. 2018 Mar 2;6(1):27. doi: 10.3390/biomedicines6010027. Biomedicines. 2018. PMID: 29498671 Free PMC article. Review.
Cited by
-
Prognosis prediction of stage IV colorectal cancer patients by mRNA transcriptional profile.Cancer Med. 2022 Dec;11(24):4900-4912. doi: 10.1002/cam4.4824. Epub 2022 May 19. Cancer Med. 2022. PMID: 35587572 Free PMC article.
-
Refining of cancer-specific genes in microsatellite-unstable colon and endometrial cancers using modified partial least square discriminant analysis.Medicine (Baltimore). 2024 Dec 27;103(52):e41134. doi: 10.1097/MD.0000000000041134. Medicine (Baltimore). 2024. PMID: 39969322 Free PMC article.
-
Identification of DNA methylation characteristics associated with metastasis and prognosis in colorectal cancer.BMC Med Genomics. 2024 May 10;17(1):127. doi: 10.1186/s12920-024-01898-4. BMC Med Genomics. 2024. PMID: 38730335 Free PMC article.
-
Construction and validation of a prognostic model based on 11 lymph node metastasis-related genes for overall survival in endometrial cancer.Cancer Med. 2022 Dec;11(23):4641-4655. doi: 10.1002/cam4.4844. Epub 2022 Jul 2. Cancer Med. 2022. PMID: 35778922 Free PMC article.
-
A Novel Glycolysis and Hypoxia Combined Gene Signature Predicts the Prognosis and Affects Immune Infiltration of Patients with Colon Cancer.Int J Gen Med. 2022 Feb 11;15:1413-1427. doi: 10.2147/IJGM.S351831. eCollection 2022. Int J Gen Med. 2022. PMID: 35185344 Free PMC article.
References
-
- Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Jin M, Frankel WL. Lymph Node Metastasis in Colorectal Cancer. Surg Oncol Clin N Am. 2018;27(2):401–12. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. - PubMed
LinkOut - more resources
Full Text Sources

