Systematic metadata analysis of brown rot fungi gene expression data reveals the genes involved in Fenton's reaction and wood decay process
- PMID: 32128279
- PMCID: PMC7033688
- DOI: 10.1080/21501203.2019.1703052
Systematic metadata analysis of brown rot fungi gene expression data reveals the genes involved in Fenton's reaction and wood decay process
Abstract
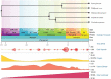
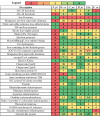
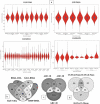
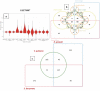
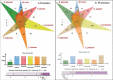
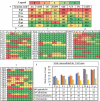
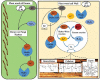
Brown-rot fungi are rapid holocellulose degraders and are the most predominant degraders of coniferous wood products in North America. Brown-rot fungi degrades wood by both enzymatic (plant biomass degrading carbohydrate active enzymes-CAZymes) and non-enzymatic systems (Fenton's reaction) mechanisms. Identifying the genes and molecular mechanisms involved in Fenton's reaction would significantly improve our understanding about brown-rot decay. Our present study identifies the common gene expression patterns involved in brown rot decay by performing metadata analysis of fungal transcriptome datasets. We have also analyzed and compared the genome-wide annotations (InterPro and CAZymes) of the selected brown rot fungi. Genes encoding for various oxidoreductases, iron homeostasis, and metabolic enzymes involved in Fenton's mechanism were found to be significantly expressed among all the brown-rot fungal datasets. Interestingly, a higher number of hemicellulases encoding genes were differentially expressed among all the datasets, while a fewer number of cellulases and peroxidases were expressed (especially haem peroxidase and chloroperoxidase). Apart from these lignocellulose degrading enzymes genes encoding for aldo/keto reductases, 2-nitro dioxygenase, aromatic-ring dioxygenase, dienelactone hydrolase, alcohol dehydrogenase, major facilitator superfamily, cytochrome-P450 monoxygenase, cytochrome b5, and short-chain dehydrogenase were common and differentially up regulated among all the analyzed brown-rot fungal datasets.
Keywords: Fenton’s reaction; Haber-Weiss reaction; Plant biomass; brown-rot fungi; lignocellulose; wood-decaying fungi.
© 2019 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Figures

Similar articles
-
Molecular Networks of Postia placenta Involved in Degradation of Lignocellulosic Biomass Revealed from Metadata Analysis of Open Access Gene Expression Data.Int J Biol Sci. 2018 Feb 9;14(3):237-252. doi: 10.7150/ijbs.22868. eCollection 2018. Int J Biol Sci. 2018. PMID: 29559843 Free PMC article.
-
Multi-omic Analyses of Extensively Decayed Pinus contorta Reveal Expression of a Diverse Array of Lignocellulose-Degrading Enzymes.Appl Environ Microbiol. 2018 Oct 1;84(20):e01133-18. doi: 10.1128/AEM.01133-18. Print 2018 Oct 15. Appl Environ Microbiol. 2018. PMID: 30097442 Free PMC article.
-
The secretome of two representative lignocellulose-decay basidiomycetes growing on sugarcane bagasse solid-state cultures.Enzyme Microb Technol. 2019 Nov;130:109370. doi: 10.1016/j.enzmictec.2019.109370. Epub 2019 Jul 3. Enzyme Microb Technol. 2019. PMID: 31421724
-
Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin.Int Microbiol. 2005 Sep;8(3):195-204. Int Microbiol. 2005. PMID: 16200498 Review.
-
Lignin-modifying enzymes in filamentous basidiomycetes--ecological, functional and phylogenetic review.J Basic Microbiol. 2010 Feb;50(1):5-20. doi: 10.1002/jobm.200900338. J Basic Microbiol. 2010. PMID: 20175122 Review.
Cited by
-
Large-scale phenotyping of 1,000 fungal strains for the degradation of non-natural, industrial compounds.Commun Biol. 2021 Jul 15;4(1):871. doi: 10.1038/s42003-021-02401-w. Commun Biol. 2021. PMID: 34267314 Free PMC article.
-
Multi-omics analysis provides insights into lignocellulosic biomass degradation by Laetiporus sulphureus ATCC 52600.Biotechnol Biofuels. 2021 Apr 17;14(1):96. doi: 10.1186/s13068-021-01945-7. Biotechnol Biofuels. 2021. PMID: 33865436 Free PMC article.
-
Transcriptome Analysis of Podoscypha petalodes Strain GGF6 Reveals the Diversity of Proteins Involved in Lignocellulose Degradation and Ligninolytic Function.Indian J Microbiol. 2022 Dec;62(4):569-582. doi: 10.1007/s12088-022-01037-6. Epub 2022 Aug 25. Indian J Microbiol. 2022. PMID: 36458217 Free PMC article.
-
Comparative transcriptomics reveals unique pine wood decay strategies in the Sparassis latifolia.Sci Rep. 2022 Nov 18;12(1):19875. doi: 10.1038/s41598-022-24171-z. Sci Rep. 2022. PMID: 36400936 Free PMC article.
-
Transcriptomic Insights into the Degradation Mechanisms of Fomitopsis pinicola and Its Host Preference for Coniferous over Broadleaf Deadwood.Microorganisms. 2025 Apr 27;13(5):1006. doi: 10.3390/microorganisms13051006. Microorganisms. 2025. PMID: 40431179 Free PMC article.
References
-
- Arantes V, Goodell B.. 2014. Current understanding of brown-rot fungal biodegradation mechanisms: a review. Deterioration and protection of sustainable biomaterials. 1158 (2014): 3–21..
-
- Bagley S, Richter D.. 2002. Biodegradation by brown rot fungi. In: Industrial applications. Springer; p. 327–341.
-
- Barb W, Baxendale J, George P, Hargrave K. 1951a. Reactions of ferrous and ferric ions with hydrogen peroxide. Part I.—the ferrous ion reaction. Trans Faraday Soc. 47:462–500.
-
- Barb W, Baxendale J, George P, Hargrave K. 1951b. Reactions of ferrous and ferric ions with hydrogen peroxide. Part II.—the ferric ion reaction. Trans Faraday Soc. 47:591–616.
-
- Barb W. G, Baxendale J. H, George P, Hargrave K. R. 1955. Reactions of ferrous and ferric ions with hydrogen peroxide. part 3.—Reactions in the presence of α: α′-dipyridyl. Trans Faraday Soc. 51:935––946..
LinkOut - more resources
Full Text Sources
Other Literature Sources
